Choose a product below to filter the page content to your needs:
Cell Ranger 6.0 and later supports analyzing 3' Cell Multiplexing data with the cellranger multi pipeline.
The cellranger multi pipeline is required to analyze 3' Cell Multiplexing data. Otherwise, users can continue to use cellranger count.
| 3' GEX | 3' FB | CellPlex | Use multi? |
|---|---|---|---|
| Yes | Yes | Yes | Required |
| Yes | Yes | No | Optional. Prefer count |
| Yes | No | Yes | Required |
| Yes | No | No | Optional. Prefer count |
| No | Yes | No | Optional. Prefer count |
First, generate FASTQ files. For example, if the flow cell ID was HAWT7ADXX, then cellranger mkfastq will output FASTQ files in HAWT7ADXX/outs/fastq_path. If you are already starting with FASTQ files, you can skip this step and proceed directly to run cellranger multi.
Running cellranger multi requires a config CSV, described below, invoking the following arguments:
| Argument | Description |
|---|---|
--id | A unique run ID string: e.g. sample345 that is also the output folder name. Cannot be more than 64 characters. |
--csv | Path to config CSV file with input libraries and analysis parameters. |
The multi config CSV contains both the library definitions and experimental design variables. It is composed of up to four sections for 3' data:
- The
[gene-expression]section has two columns that specify parameters relevant to analysis of gene expression data, such as reference genome and cell-calling parameters, as well as other all-purpose parameters. - The
[feature]section has two columns that specify parameters relevant to analysis of Feature Barcode libraries. - The
[libraries]section has three required columns that specify where the input FASTQ files may be found. - The
[samples]section has two required columns that specify sample information for Cell Multiplexing.
Go to the Cell Ranger Multi Config CSV page for a complete list of options for each section.
Generate a multi config CSV template by running cellranger multi-template, see usage here.
Example formats for different product configurations are below. Example multi config CSVs can be downloaded from Single Cell Gene Expression with Cell Multiplexing public datasets.
After determining these input arguments, run cellranger multi (replace code in red with relevant file path and ID name):
cd /home/jdoe/runs
cellranger multi --id=sample345 --csv=/home/jdoe/sample345.csv
Following a series of checks to validate input arguments, cellranger multi pipeline stages will begin to run:
Martian Runtime - v4.0.8
Running preflight checks (please wait)...
...
By default, Cell Ranger will use all of the cores available on your system to execute pipeline stages. You can specify a different number of cores to use with the --localcores option; for example, --localcores=16 will limit Cell Ranger to using up to sixteen cores at once. Similarly, --localmem will restrict the amount of memory (in GB) used by Cell Ranger.
The pipeline will create a new folder named with the run ID you specified using the --id argument (e.g. /home/jdoe/runs/sample345) for its output. If this folder already exists, Cell Ranger will assume it is an existing pipestance and attempt to resume running it.
A successful cellranger multi run should conclude with a message similar to this:
Waiting 6 seconds for UI to do final refresh.
Pipestance completed successfully!
yyyy-mm-dd hh:mm:ss Shutting down.
Saving pipestance info to "tiny/tiny.mri.tgz"
The outputs of the pipeline will be contained in a folder named with the run ID you specified (e.g. sample345). The subfolder named outs/ will contain the main pipeline outputs.
Here are a few example multi config CSVs for some common product configurations, along with simplified diagrams for the corresponding experimental set up. Make sure to replace /path/to with the absolute path to your data, and edit the necessary text according to the experiment's sample/library/file names.
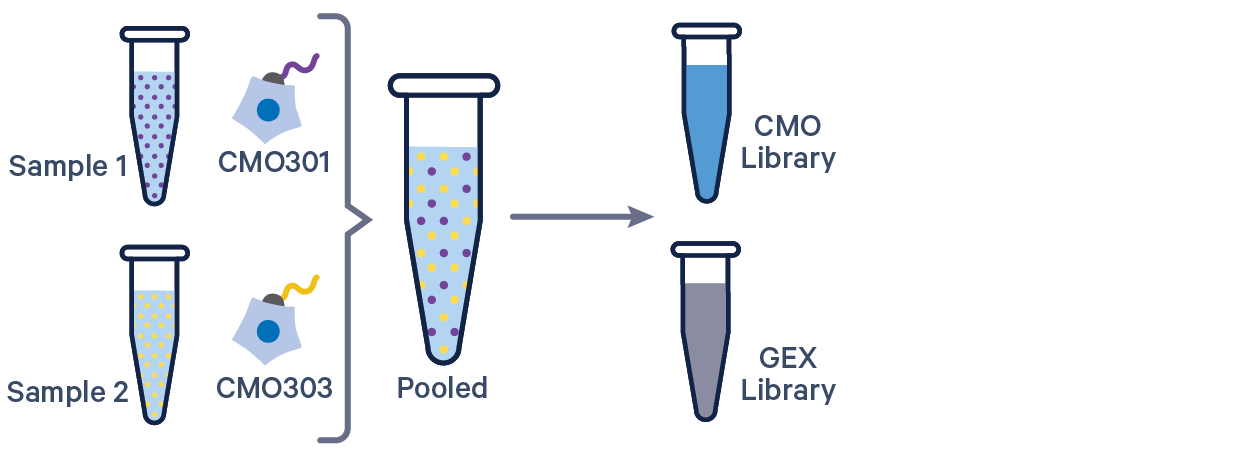
[gene-expression]
reference,/path/to/transcriptome
[libraries]
fastq_id,fastqs,feature_types
gex1,/path/to/fastqs,Gene Expression
mux1,/path/to/fastqs,Multiplexing Capture
[samples]
sample_id,cmo_ids
sample1,CMO301
sample2,CMO303
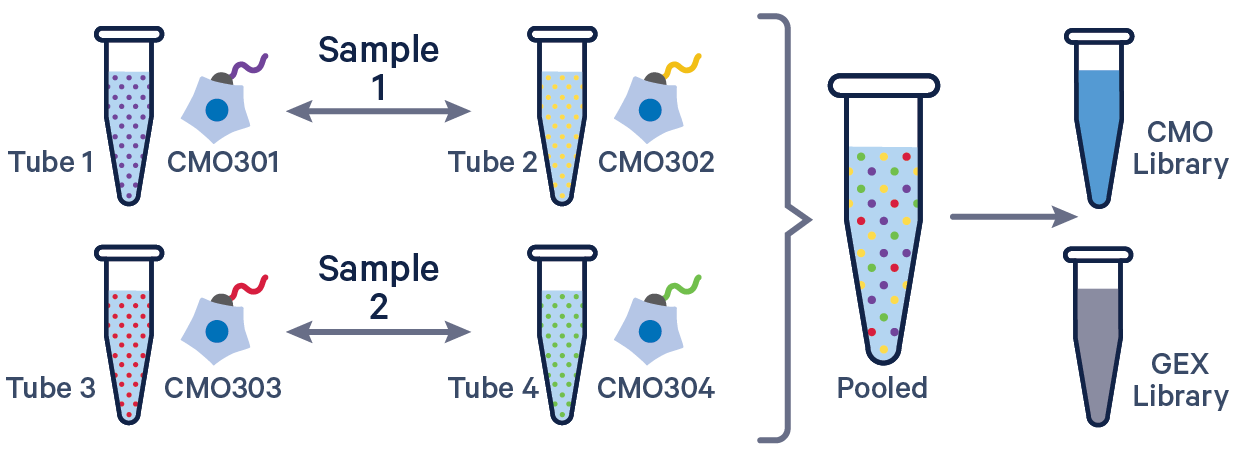
See example dataset. Note usage of the | to separate CMO tags. Learn more about when to use multiple CMOs per sample here.
[gene-expression]
reference,/path/to/transcriptome
[libraries]
fastq_id,fastqs,feature_types
gex1,/path/to/fastqs,Gene Expression
mux1,/path/to/fastqs,Multiplexing Capture
[samples]
sample_id,cmo_ids
sample1,CMO301|CMO302
sample2,CMO303|CMO304
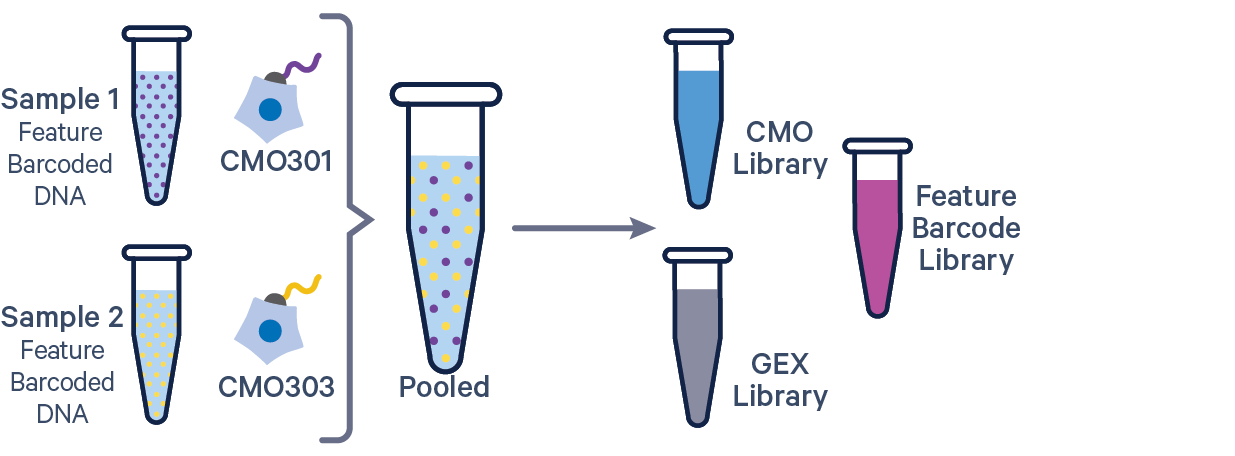
The additional Feature Barcode library in this config CSV example ([libraries] section) is Antibody Capture. Specify CRISPR Guide Capture as the feature_types for CRISPR Feature Barcode experiments.
[gene-expression]
reference,/path/to/transcriptome
[feature]
reference,/path/to/feature_reference.csv
[libraries]
fastq_id,fastqs,feature_types
gex1,/path/to/fastqs,Gene Expression
abc1,/path/to/fastqs,Antibody Capture
mux1,/path/to/fastqs,Multiplexing Capture
[samples]
sample_id,cmo_ids
sample1,CMO301
sample2,CMO303
The cmo-set option in the [gene-expression] section of the multi config CSV allows you to provide a reference for custom Cell Multiplexing oligos (e.g., antibody TotalSeqA/B/C tags). The design of this reference is nearly identical to the Feature Barcode Reference used to describe Feature Barcodes, with one difference: the feature_type is required to be Multiplexing Capture instead of those feature types supported in the Feature Barcode reference. The id column may contain alphanumeric, underscore, and hyphen characters; special characters like a pipe (|) should not be used in this file (only for separating multiple CMO IDs from the same sample in config CSV).
For example, Cell Ranger's default CMO reference looks like this (built into Cell Ranger):
id,name,read,pattern,sequence,feature_type
CMO301,CMO301,R2,5P(BC),ATGAGGAATTCCTGC,Multiplexing Capture
CMO302,CMO302,R2,5P(BC),CATGCCAATAGAGCG,Multiplexing Capture
CMO303,CMO303,R2,5P(BC),CCGTCGTCCAAGCAT,Multiplexing Capture
CMO304,CMO304,R2,5P(BC),AACGTTAATCACTCA,Multiplexing Capture
CMO305,CMO305,R2,5P(BC),CGCGATATGGTCGGA,Multiplexing Capture
CMO306,CMO306,R2,5P(BC),AAGATGAGGTCTGTG,Multiplexing Capture
CMO307,CMO307,R2,5P(BC),AAGCTCGTTGGAAGA,Multiplexing Capture
CMO308,CMO308,R2,5P(BC),CGGATTCCACATCAT,Multiplexing Capture
CMO309,CMO309,R2,5P(BC),GTTGATCTATAACAG,Multiplexing Capture
CMO310,CMO310,R2,5P(BC),GCAGGAGGTATCAAT,Multiplexing Capture
CMO311,CMO311,R2,5P(BC),GAATCGTGATTCTTC,Multiplexing Capture
CMO312,CMO312,R2,5P(BC),ACATGGTCAACGCTG,Multiplexing Capture
The default CMO reference above is available as a downloadable CSV.
The barcode-sample-assignment option in the [gene-expression] section of the multi config CSV allows users to provide a file that manually specifies the barcodes for each sample. It will override Cell Ranger's default cell calling and tag calling steps, and may be useful in cases where data with microfluidic failures can be partially rescued. This feature allows users to import custom tag calling done via 3rd party tools as well (see the Tag assignment of 10x Genomics CellPlex data using Seurat's HTODemux function Analysis Guide for help).
Here is an example multi config CSV:
[gene-expression]
reference,/path/to/transcriptome
barcode-sample-assignment,/path/to/barcode_sample_assignment.csv
[libraries]
fastq_id,fastqs,feature_types
gex1,/path/to/fastqs,Gene Expression
mux1,/path/to/fastqs,Multiplexing Capture
[samples]
sample_id,cmo_ids
sample1,CMO301
sample2,CMO303
The barcode-sample CSV file has at most two columns, one for the barcode sequence and another that is either the sample ID or the tag assignment. A barcode can only be assigned to one sample; barcodes with multiple sample or tag entries will result in an error in Cell Ranger. Here are two examples:
Option 1: Assign to samples
Barcode,Sample_ID
ACGTACGTACGTACGT-1,Jurkat
CGTACGTACGTACGTA-1,Raji
GTACGTACGTACGTAC-1,Jurkat
TACGTACGTACGTACG-1,Raji
...
Option 2: Assign to tags
Barcode,Assignment
ACGTACGTACGTACGT-1,CMO1
CGTACGTACGTACGTA-1,Multiplet
GTACGTACGTACGTAC-1,Blank
TACGTACGTACGTACG-1,Unassigned
...