Instructions for building the 10x Genomics Public Reference are documented separately.
2024-A reference packages are not backward compatible with Cell Ranger v5.0.1 and prior.
- Human GRCh38 (GENCODE v44/Ensembl110 annotations)
- Mouse GRCm39 (GENCODE vM33/Ensembl110 annotations)
- Human and mouse GRCh38 and GRCm39
Update notes:
- Human transcriptome annotations have been updated from GENCODE v32 to GENCODE v44.
- Mouse transcriptome annotations have been updated from vM23 to vM33.
- Readthrough annotations have been improved, and erroneous systematic gene names have been removed.
- Polymorphic pseudogenes were included by adding
protein_coding_LoFto the list of accepted biotypes. - Pseudoautosomal regions (PAR) on the Y chromosome have been masked and the pseudoautosomal genes on the Y chromosome have been removed from the GTF. The corresponding genes on the X chromosome are still present in the GTF and will have associated counts.
- Summary of changes is shown in the table:
| Human | Mouse | |
|---|---|---|
| Number of new gene IDs | 2339 | 1746 |
| Number of genes removed | 301 | 335 |
| Number of gene names changed | 12913 | 1905 |
| Number of gene IDs changed (based on gene name) | 69 | 56 |
2020-A reference packages are backward compatible with Cell Ranger v3.1.0 and prior.
Human GRCh38 (GENCODE v32/Ensembl98)
Mouse mm10 (GENCODE vM23/Ensembl98)
Human and mouse GRCh38 and mm10
Update notes:
- Transcriptome annotations updated from Ensembl 93 to GENCODE v32 (human) and vM23 (mouse), which are equivalent to Ensembl 98.
- GRCh38 and mm10 sequences are not changed; chromosome names now follow the GENCODE/UCSC convention (e.g., chr1 and chrM) rather than the Ensembl convention (1 and MT).
- Additional filtering removes genes with unreliable annotations that often overlap more legitimate genes (see build scripts for details), resulting in improved overall sensitivity.
- Mapping rates and gene/UMI sensitivity are increased due to more comprehensive annotations and improved manual curation of genes:
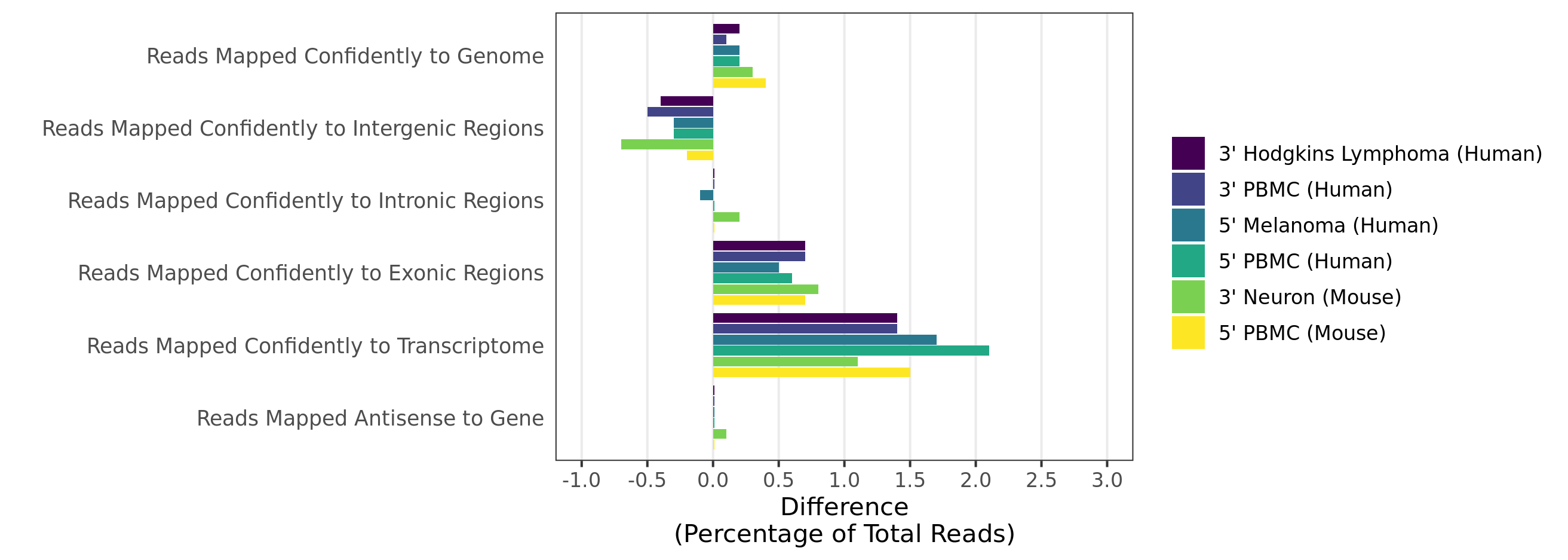
Human and mouse 3.1.0 GRCh38 and mm10
Human 3.0.0 GRCh38 Human 3.0.0 hg19 Mouse 3.0.0 mm10 Human and mouse 3.0.0 hg19 and mm10
Mouse mm10 (V(D)J genes included) Human and house hg19 and mm10 (with V(D)J)
Human 1.2.0 GRCh38 Human 1.2.0 hg19 Mouse 1.2.0 mm10 Human and mouse 1.2.0 hg19 and mm10 ERCC reference ercc92
Human V(D)J reference GRCh38
The Human V(D)J reference has been updated to exclude the following genes:
- IGHV4-30-2
- IGKV1D-33
- IGKV1D-37
- IGKV1D-39
- IGKV2D-28
These genes have counterparts with identical V, D, J, and C gene sequences, but differ in the length of their 5' UTRs. Removing duplicates improves clonotype assignment.
- Added human gene IGHV3-9
- For two genes that are identical except for extra bases on the 3' end, only the longer version was retained. List of affected genes:
HUMAN:
IGHA1 ENST00000390547
IGHD ENST00000390556
IGHD1-1 ENST00000454908
IGHD1-14 ENST00000451044
IGHD1-20 ENST00000450276
IGHD1-26 ENST00000390567
IGHD1-7 ENST00000430425
IGHD1/OR15-1A ENST00000605284
IGHD2-15 ENST00000390578
IGHD2-2 ENST00000390591
IGHD2-21 ENST00000390572
IGHD2-8 ENST00000390585
IGHD2/OR15-2A ENST00000603077
IGHD3-10 ENST00000390583
IGHD3-16 ENST00000390577
IGHD3-22 ENST00000390571
IGHD3-3 ENST00000390590
IGHD3-9 ENST00000390584
IGHD3/OR15-3A ENST00000604950
IGHD4-11 ENST00000431440
IGHD4-17 ENST00000431870
IGHD4-23 ENST00000437320
IGHD4/OR15-4A ENST00000603326
IGHD5-12 ENST00000390581
IGHD5-18 ENST00000390575
IGHD5-24 ENST00000390569
IGHD5/OR15-5A ENST00000604642
IGHD6-13 ENST00000390580
IGHD6-19 ENST00000390574
IGHD6-25 ENST00000452198
IGHD6-6 ENST00000454691
IGHD7-27 ENST00000439842
IGHG1 ENST00000390542
IGHG1 ENST00000390548
IGHG1 ENST00000390549
IGHG2 ENST00000390545
IGHG3 ENST00000390551
IGHG4 ENST00000390543
IGHJ1 ENST00000390565
IGHM ENST00000390559
IGHV1-18 ENST00000390605
IGHV1-2 ENST00000390594
IGHV1-24 ENST00000390610
IGHV1-3 ENST00000390595
IGHV1-45 ENST00000390621
IGHV1-46 ENST00000390622
IGHV1-58 ENST00000390628
IGHV1-69 ENST00000390633
IGHV1-69-2 ENST00000615784
IGHV2-26 ENST00000390611
IGHV2-5 ENST00000390597
IGHV2-70D ENST00000390634
IGHV3-11 ENST00000390601
IGHV3-13 ENST00000390602
IGHV3-15 ENST00000390603
IGHV3-16 ENST00000390604
IGHV3-20 ENST00000390606
IGHV3-21 ENST00000390607
IGHV3-23 ENST00000390609
IGHV3-30 ENST00000603660
IGHV3-35 ENST00000390617
IGHV3-38 ENST00000390618
IGHV3-43 ENST00000434710
IGHV3-48 ENST00000390624
IGHV3-49 ENST00000390625
IGHV3-53 ENST00000390627
IGHV3-64 ENST00000454421
IGHV3-66 ENST00000390632
IGHV3-7 ENST00000390598
IGHV3-72 ENST00000433072
IGHV3-73 ENST00000390636
IGHV3-74 ENST00000424969
IGHV4-28 ENST00000390612
IGHV4-34 ENST00000390616
IGHV4-39 ENST00000390619
IGHV4-4 ENST00000455737
IGHV4-59 ENST00000390629
IGHV4-61 ENST00000390630
IGHV5-51 ENST00000390626
IGHV6-1 ENST00000390593
IGKV1-12 ENST00000480492
IGKV1-16 ENST00000479981
IGKV1-17 ENST00000490686
IGKV1-27 ENST00000498435
IGKV1-33 ENST00000473726
IGKV1-37 ENST00000465170
IGKV1-39 ENST00000498574
IGKV1-5 ENST00000496168
IGKV1-6 ENST00000464162
IGKV1-8 ENST00000495489
IGKV1-9 ENST00000493819
IGKV2-24 ENST00000484817
IGKV2-28 ENST00000482769
IGKV2-30 ENST00000468494
IGKV3-11 ENST00000483158
IGKV3-15 ENST00000390252
IGKV3-20 ENST00000492167
IGKV3-7 ENST00000390247
IGKV3D-7 ENST00000443397
IGKV5-2 ENST00000390244
IGKV6-21 ENST00000390256
IGLV1-36 ENST00000390301
IGLV1-40 ENST00000390299
IGLV1-44 ENST00000628287
IGLV2-33 ENST00000390302
IGLV3-32 ENST00000390303
IGLV5-37 ENST00000390300
IGLV7-43 ENST00000390298
TRBD1 ENST00000631435
TRBJ1-1 ENST00000634213
TRBJ1-2 ENST00000631745
TRBJ1-3 ENST00000633780
TRBJ1-4 ENST00000632041
TRBJ1-5 ENST00000634000
TRBJ2-1 ENST00000390412
TRBJ2-2 ENST00000390413
TRBJ2-2P ENST00000390414
TRBJ2-3 ENST00000390415
TRBJ2-4 ENST00000390416
TRBJ2-5 ENST00000390417
TRBJ2-6 ENST00000390418
TRBV10-1 ENST00000390364
TRBV11-1 ENST00000390367
TRBV11-3 ENST00000611787
TRBV12-3 ENST00000620569
TRBV13 ENST00000614171
TRBV14 ENST00000617639
TRBV15 ENST00000616518
TRBV16 ENST00000620773
TRBV23-1 ENST00000390396
TRBV27 ENST00000390399
TRBV28 ENST00000390400
TRBV29-1 ENST00000422143
TRBV3-1 ENST00000390387
TRBV4-2 ENST00000390392
TRBV5-1 ENST00000390381
TRBV5-6 ENST00000390375
TRBV5-7 ENST00000390378
TRBV6-1 ENST00000390353
TRBV6-5 ENST00000390368
TRBV7-1 ENST00000547918
TRBV7-7 ENST00000390377
TRGJ1 ENST00000390337
MOUSE:
Added missing mouse TRGV and TRGC genes
TRGC1 ENSMUST00000103558 TRGC2 ENSMUST00000103561 TRGC3 ENSMUST00000198163 TRGC4 ENSMUST00000179181 TRGV1 ENSMUST00000103564 TRGV3 ENSMUST00000198663 TRGV4 ENSMUST00000103554 TRGV5 ENSMUST00000199017 TRGV6 ENSMUST00000198330 TRGV7 ENSMUST00000103553
For two genes that are identical except for extra bases on the 3' end, only the longer version was retained. List of affected genes:
IGHD2-5 ENSMUST00000178549 IGHD5-2 ENSMUST00000179166 TRAV11D ENSMUST00000103648 TRAV12D-1 ENSMUST00000181360 TRAV12D-2 ENSMUST00000197007 TRAV13D-2 ENSMUST00000197954 TRAV14D-1 ENSMUST00000181038 TRAV14D-2 ENSMUST00000196802 TRAV15D-2-DV6D-2 ENSMUST00000199800 TRAV3D-3 ENSMUST00000196023 TRAV4D-3 ENSMUST00000103592 TRAV4D-4 ENSMUST00000103600 TRAV5D-4 ENSMUST00000179701 TRAV6-6 ENSMUST00000103584 TRAV7-2 ENSMUST00000103636 TRAV7D-5
The recommended V(D)J reference packages for human and mouse have been updated from v4.0-5.0. The changes to the V(D)J reference sequences are listed below:
HUMAN:
Replace IGKV2D-40, whose leader sequence appears to be truncated.
Delete IGKV2-18, which is probably a pseudogene.
Delete IGLV5-48, which is truncated on the right.
Delete TRBV21-1, which has multiple frameshifts.
Add IGHV4-30-4, which was missing.
Add IGKV1-NL1, which was missing.
Add IGHV4-38-2, which was missing.
MOUSE:
Delete TRAV23, which is frame-shifted.
Delete the first base of the constant region gene IGHG2B.
Make a six-base insertion in IGKV12-89, based on empirical data.
Correct IGHV8-9, whose amino acid sequence showed the canonical C at the end of FWR3 as S. This is consistent with 10x data.
Add an allele of IGKV2-109, which was missing.
Add IGKV4-56, which was missing.
Add IGHV1-2, which was missing.
Recommended V(D)J reference packages for human and mouse have been updated from version 3.1.0 to 4.0.0. The changes to the V(D)J reference sequences are listed below:
- Remove the first base of the C region in certain cases. In these cases we observe that in most transcripts, the J region and C region overlap by exactly one base.
- Add an allele of the gene IGHJ6 to the human V(D)J reference.
Updates to prebuilt reference: https://support.10xgenomics.com/single-cell-vdj/software/pipelines/3.1/advanced/built-in-refs