Visium slides have features that Space Ranger users should be familiar with, particularly the slide serial number and the capture area associated with each sample. There are currently four types of slides available:
| Visium Slides |
|---|
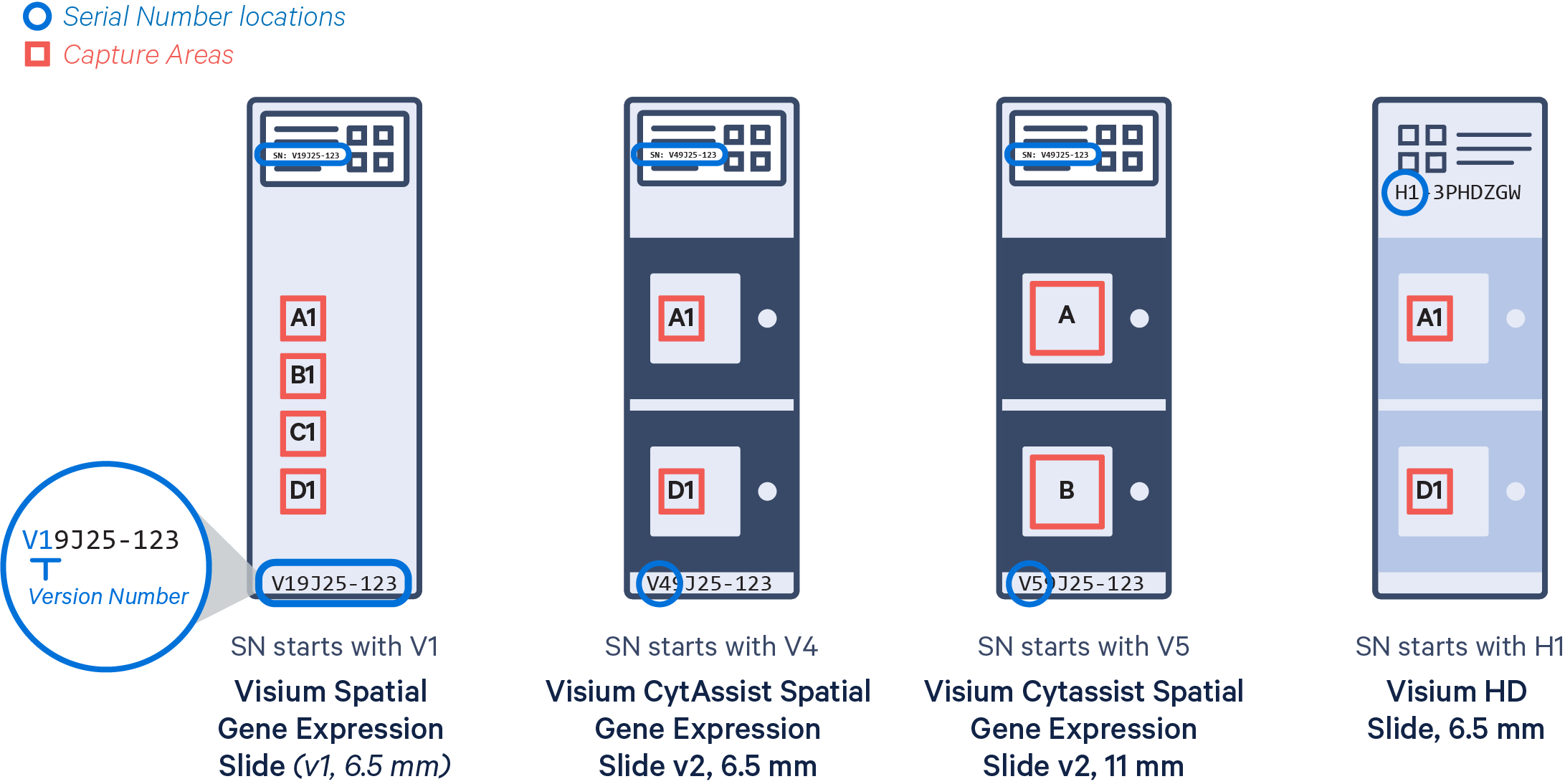 |
- Visium HD is specific to assays that use the Visium HD slide (6.5 mm). Only compatible with the Visium CytAssist instrument.
- Visium v2 includes assays that use the Visium CytAssist Spatial Gene Expression slides (v2, 6.5 mm and v2, 11 mm). Only compatible with the Visium CytAssist instrument.
- Visium v1 refers to direct placement assays that use Visium Spatial Gene Expression slides (v1, 6.5 mm).
| Capture Areas |
|---|
 |
Alignment File: A JSON file produced by Loupe Browser while using manual alignment.
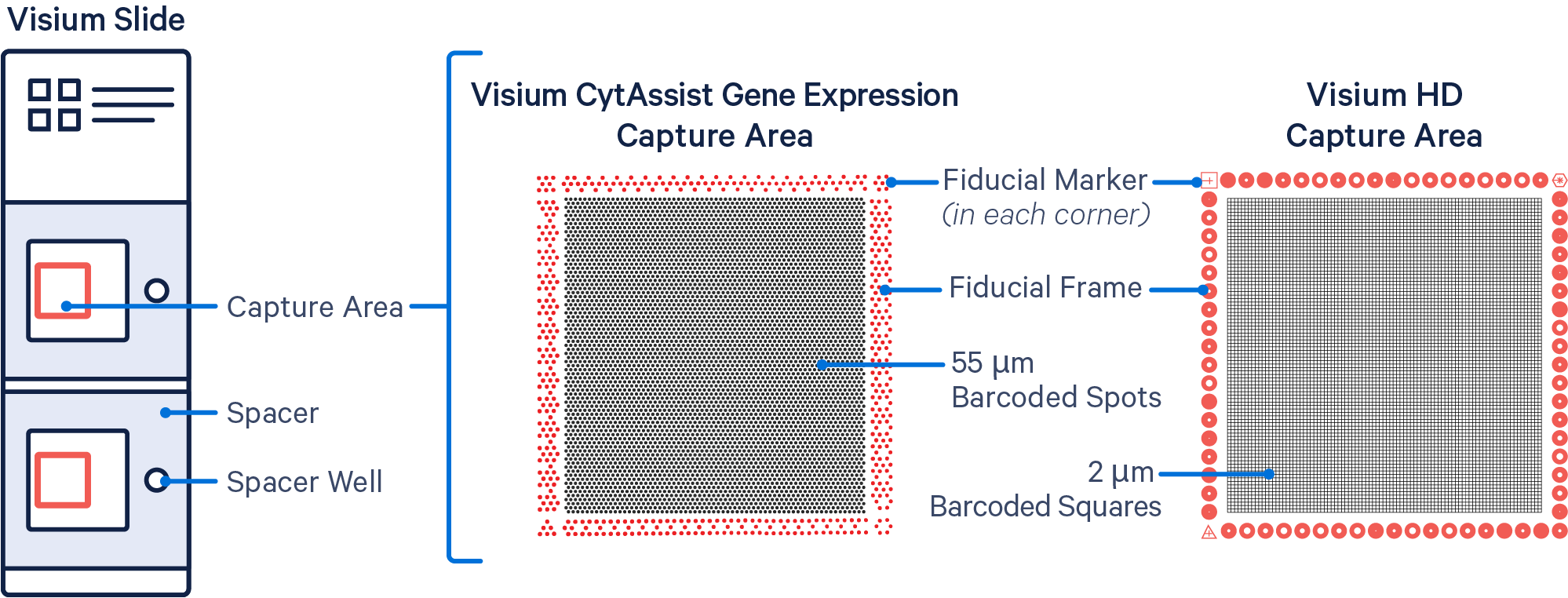
Barcoded Spots / Squares: The invisible features on the slide that contain specialized oligos for capturing poly-adenylated mRNA, probes for Gene Expression, and/or antibody oligo tags for Protein Expression, depending on the Visium assay. Visium HD capture areas contain a continuous grid of 2x2 µm barcoded squares, ~11.2 million in total. By contrast, Visium v1 and v2 capture areas have an array of ~5,000 barcoded spots, 55 µm in diameter, spaced 100 µm apart. See also: Spatial Barcodes.
Brightfield Image: A light-microscopy image depicting tissue on a slide. In a Visium experiment, a brightfield image is used as an anatomical reference. These images are typically stained with hematoxylin and eosin (H&E) to highlight tissue structure. See also: Microscope Image.
Capture Area: Active regions for capturing expression data on a Visium slide. Slides either have two or four capture areas. Slide areas are named consecutively from top to bottom:
- A1, B1, C1, D1 for Visium Spatial Gene Expression slides, v1, 6.5 mm
- A, B for Visium CytAssist Spatial Gene Expression slide, v2, 11 mm
- A1, D1 for Visium CytAssist Spatial Gene Expression slides, v2, 6.5 mm, and Visium HD slides, 6.5 mm.
Direct Placement: Visium assays where the tissue is placed directly on the Visium slide, without the use of the Visium CytAssist instrument.
Dual Indexing: A strategy for sequencing multiple samples on the same flow cell by using two oligonucleotide sequences, one attached to either end of each fragment to be sequenced, in order to uniquely identify the sample. Visium libraries require dual-indexing. See also: Sample Index.
Feature: A unique type of countable molecule. Can refer to a gene or a Spatial Barcode Antibody. Each feature is either a gene declared in the transcriptome reference or an antibody declared in the feature reference file. Corresponds to a row in the Feature-barcode Matrix.
Feature-barcode Matrix: A matrix of counts representing the number of unique observations of each feature for each spatial barcode. Genes are defined by the reference, and feature barcodes defined in the feature reference appear as rows in the matrix. Each spatial barcode is a column of the matrix.
Feature Reference: A CSV file declaring the name, read layout, and barcode sequence of all the spatial barcode antibody reagents in use in an experiment. A feature_reference.csv must be provided to spaceranger count when analyzing protein expression data.
Fiducial Frame: A frame of specially patterned spots surrounding each capture area on a Visium slide. These spots are used by Space Ranger to determine where the capture area is in an image.
Fiducial Marker: A subset of the fiducial spots in each corner of the capture area on a Visium slide. Fiducial markers are easily identifiable shapes (hourglass, triangle, open hexagon, and filled hexagon).
Fluorescence Imaging: An imaging technique whereby signal is generated by fluorophores that re-emit narrow-spectrum light when excited by light of a certain wavelength or color.
Gene Expression (GEX): A process by which the information encoded in a gene is transcribed to mRNA molecules.
H&E Staining: The process of applying hematoxylin and eosin to tissue in order to highlight tissue structure. Hematoxylin colors cell nuclei blue, and eosin colors the cytoplasm and extracellular matrix pink.
Immunohistochemistry (IHC) and Immunofluorescence (IF): IHC is an imaging technique whereby protein is stained using a matching, antibody-based label. Immunofluorescence (IF) is a subset of this technique that generates a signal using fluorophores attached to antibodies and fluorescence imaging.
Library (or Sequencing Library): A Visium library prepared from a single capture area.
Microscope Image: A high-resolution brightfield or fluorescence image of the tissue section on the standard glass slide captured by a microscope.
Probe Filter: Within the whole-transcriptome Probe Set reference file declaring the gene panel used for a Visium for FFPE experiment there is an included column. By default, probes predicted to have some off-target activity to homologous genes or sequences are excluded from analysis. Users can include UMI counts from all probes, including those with potential off-target activity, with the --filter-probes=false option.
Probe Set: A whole-transcriptome reference file declaring the gene panel used for a Visium for FFPE experiment, which specifies detailed information about the genes targeted by each probe. This file must be provided to spaceranger count with the --probe-set option when analyzing Visium for FFPE data.
Protein Expression: a feature type supported by Visium CytAssist (see also Spatial Barcode Antibody).
RNA-Templated Ligation: Visium for FFPE is designed around a strict probe pairing framework. For each target locus, when two half-probe sequences bind to the proper locus and ligate together during the assay, a countable barcode-UMI-probe product is made.
Sample: A single tissue section analyzed.
Sample Index: An oligonucleotide sequence used in library construction to differentiate multiple samples that are sequenced on the same flow cell. On the Illumina platform, these sequences are read out as separate index reads and reads are sorted into sample-specific files by the demultiplexing software (mkfastq, bcl2fastq, or bcl-convert). Visium libraries require dual-indexing.
Sequencing Run: The output data, including Illumina BCL files, from one sequencing instrument run. The data can be demultiplexed by lane or by sample index.
Slide File: A file describing the layout of capture spots in a single slide identified by slide serial number. Can be queried by serial number from the Download Center.
Slide Serial Number: The unique identifier printed on the label of each Visium slide. The serial number starts with a prefix indicating the slide type:
- V1 = Visium Spatial Gene Expression Slide (v1, 6.5 mm)
- V4 = Visium CytAssist Spatial Gene Expression Slide (v2, 6.5 mm)
- V5 = Visium CytAssist Spatial Gene Expression Slide (v2, 11 mm)
- H1 = Visium HD Slide (6.5 mm)
Spatial Barcode: The subsequence of a Spatial Barcode read that uniquely identifies the feature (spots for Visium v1/v2 assays and squares for Visium HD).
Spatial Barcode Antibody (or Antibody): A reagent consisting of an antibody with high affinity to a known protein conjugated to an Antibody Barcode oligonucleotide that identifies the antibody. These reagents are used to detect the presence of proteins. Supported only with the Visium CytAssist Spatial Gene and Protein Expression product.
Visium CytAssist: An instrument that mediates the tissue permeabilization to release the probes (and antibody tags) from tissues on a standard glass slide for capture by spatially barcoded oligonucleotides within each capture area on the Visium slide. It also captures the image of the tissue section aligned with capture areas on the Visium CytAssist slide. See also: Visium CytAssist-captured image.
Visium CytAssist-captured image (or CytAssist Image): A brightfield image of the eosin stained tissue section in TIFF format captured by the Visium CytAssist instrument. This image contains the fiducial frame.