On this page, we describe how to format custom sequence information for common Xenium v1 and Xenium Prime advanced custom targets. Please send the files associated with these targets to your 10x bioinformatics representative.
This is part of the advanced workflow to request species standalone or advanced custom panels. Guidance is provided below for the following advanced targets:
- Single nucleotide variants
- Indels
- Gene isoforms
- Exogenous genes
- T cell receptors
- Barcode detection
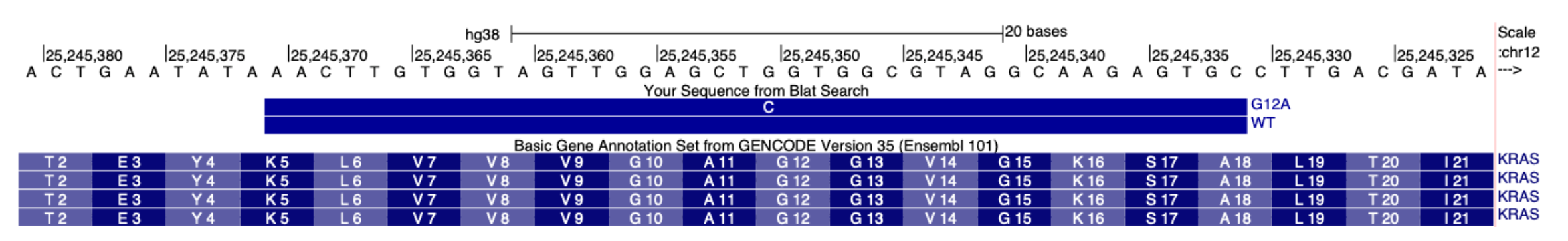
SNV performance will vary due to the ligation junction preferences of the ligase used in the Xenium In Situ assay workflow. Preferred ligation junctions are described in the Species Standalone Custom and Advanced Custom Panel Design for Xenium In Situ Technical Note. Provide a FASTA file with exonic sequences. Here is an example FASTA file for specifying the KRAS G12D mutation:
>NM_004985 KRAS-CDS-sequence
ATGACTGAATATAAACTTGTGGTAGTTGGAGCTGGTGGCGTAGGCAAGAGTGCCTTGACGATACAGCTAA
TTCAGAATCATTTTGTGGACGAATATGATCCAACAATAGAGGATTCCTACAGGAAGCAAGTAGTAATTGA
TGGAGAAACCTGTCTCTTGGATATTCTCGACACAGCAGGTCAAGAGGAGTACAGTGCAATGAGGGACCAG
TACATGAGGACTGGGGAGGGCTTTCTTTGTGTATTTGCCATAAATAATACTAAATCATTTGAAGATATTC
ACCATTATAGAGAACAAATTAAAAGAGTTAAGGACTCTGAAGATGTACCTATGGTCCTAGTAGGAAATAA
ATGTGATTTGCCTTCTAGAACAGTAGACACAAAACAGGCTCAGGACTTAGCAAGAAGTTATGGAATTCCT
TTTATTGAAACATCAGCAAAGACAAGACAGGGTGTTGATGATGCCTTCTATACATTAGTTCGAGAAATTC
GAAAACATAAAGAAAAGATGAGCAAAGATGGTAAAAAGAAGAAAAAGAAGTCAAAGACAAAGTGTGTAAT
TATGTAA
Additionally, provide a separate CSV file describing the single nucleotide variants you would like to target. This file should use 0-based coordinates in the position (pos) column, with each alternative base in the alt column (one row per alternative base). In the example below, pos = 34 indicates the 35th base of the sequence above in a 0-based coordinates system, where the SNV is an A instead of a G:
sequence,pos,alt
NM_004985,34,A
An example of the designed probes aligned to the human GRCh38 reference transcriptome is shown below:
Provide a FASTA file with exonic sequence. Here is an example:
>indel
ATGCATGCATGCATGCATGCATGCATGCATGCATGCATGCGGTCTCGATGTTGTCAATATTCCCCCAAGAACCCTTCTGGACAATGCATGCATGCATGCATGCATGCATGCATGCATGCATGC
Additionally, provide a CSV file to define insertions and deletions. The file should have four columns: sequence, start, end, and alt. The alternative sequence should be left-padded and the positions should be 0-based. For example, the CSV file should look like this:
sequence,start,end,alt
indel,42,43,TAA
indel,41,43,
indel,21,51,
Here are three scenarios illustrating how 10x will design the probes for Xenium v1 using the example FASTA and CSV above:
-
Short insertion: This adds an AA at position 42. We left-pad the sequence with TAA, which results in this design:
seq: GCATGCATGCATGCATGCGGT CTCGATGTTGTCAATATTCC WT probe: GCATGCATGCATGCATGCGGT CTCGATGTTGTCAATATTC ALT probe: CATGCATGCATGCATGCGGTAACTCGATGTTGTCAATATT -
Short deletion: This deletes a GT. The
altcolumn is empty.seq: TGCATGCATGCATGCATGCGGTCTCGATGTTGTCAATATTCC WT probe: CATGCATGCATGCATGCGGTCTCGATGTTGTCAATATTCC ALT probe: TGCATGCATGCATGCATGCG CTCGATGTTGTCAATATTCC -
Large deletion: This removes 30 bp (upper case represents the deletion region).
seq: agagagccttgaggaaaaccaGCGGAACCTCCTTCAGATGACTGAAAAGTTcttccatgccatcatcagttc WT probe: gagagccttgaggaaaaccaGCGGAACCTCCTTCAGATGA ALT probe: gagagccttgaggaaaacca cttccatgccatcatcagtt
Please supply a FASTA file with the sequence of each splice junction you want to target, where the splice junction is centered and there are at least 40 bases of transcribed sequence on both sides in transcription orientation.
Here is an example FASTA file for specifying probes for the human EGFRvIII variant, where exon 1 aberrantly splices to exon 8.
>EGFR_exon1_exon8
GCGCTCTGCCCGGCGAGTCGGGCTCTGGAGGAAAAGAAAGGTAATTATGTGGTGACAGATCACGGCTCGTGCGTCCGAGC
>EGFR_exon1_exon2
GCGCTCTGCCCGGCGAGTCGGGCTCTGGAGGAAAAGAAAGTTTGCCAAGGCACGAGTAACAAGCTCACGCAGTTGGGCAC
These sequences are 80 bp in total length with the splice junction in the center.
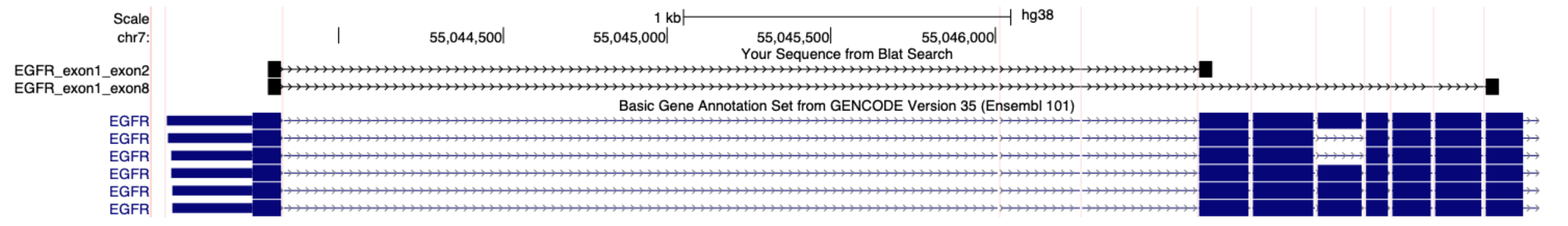
The above FASTA file is aligned to the human GRCh38 reference transcriptome. Each sequence is evenly split over the target splice junctions. Note that the sequences are in transcription orientation and only contain exonic sequence.
Exogenous genes include protein tags, fluorescent reporters, transgenes, or expressed sequences such as CRISPR guides. All of these genes can be specified by providing a FASTA file of the sequence you would like to target. Here is an example FASTA file for GFP:
>L29345.1 Aequorea victoria green-fluorescent protein (GFP) mRNA, complete cds
TACACACGAATAAAAGATAACAAAGATGAGTAAAGGAGAAGAACTTTTCACTGGAGTTGTCCCAATTCTT
GTTGAATTAGATGGCGATGTTAATGGGCAAAAATTCTCTGTCAGTGGAGAGGGTGAAGGTGATGCAACAT
ACGGAAAACTTACCCTTAAATTTATTTGCACTACTGGGAAGCTACCTGTTCCATGGCCAACACTTGTCAC
TACTTTCTCTTATGGTGTTCAATGCTTTTCAAGATACCCAGATCATATGAAACAGCATGACTTTTTCAAG
AGTGCCATGCCCGAAGGTTATGTACAGGAAAGAACTATATTTTACAAAGATGACGGGAACTACAAGACAC
GTGCTGAAGTCAAGTTTGAAGGTGATACCCTTGTTAATAGAATCGAGTTAAAAGGTATTGATTTTAAAGA
AGATGGAAACATTCTTGGACACAAAATGGAATACAACTATAACTCACATAATGTATACATCATGGCAGAC
AAACCAAAGAATGGAATCAAAGTTAACTTCAAAATTAGACACAACATTAAAGATGGAAGCGTTCAATTAG
CAGACCATTATCAACAAAATACTCCAATTGGCGATGGCCCTGTCCTTTTACCAGACAACCATTACCTGTC
CACACAATCTGCCCTTTCCAAAGATCCCAACGAAAAGAGAGATCACATGATCCTTCTTGAGTTTGTAACA
GCTGCTGGGATTACACATGGCATGGATGAACTATACAAATAAATGTCCAGACTTCCAATTGACACTAAAG
TGTCCGAACAATTACTAAATTCTCAGGGTTCCTGGTTAAATTCAGGCTGAGACTTTATTTATATATTTAT
AGATTCATTAAAATTTTATGAATAATTTATTGATGTTATTAATAGGGGCTATTTTCTTATTAAATAGGCT
ACTGGAGTGTAT
T cell receptor (TCR) detection in Xenium In Situ can come in one of two general flavors: 1) detection of Complementarity-Determining Region 3 (CDR3) sequences or 2) unbiased profiling of V/J segments. Unbiased V/J profiling is actively being developed by 10x Genomics, but is not currently available.
CDR3 sequence detection relies on using an assembled CDR3 sequence to design probes specific for the target CDR3 (such as one produced by the 10x Immune Profiling solution). By doing this, we make the assumption that there are no other CDR3 present at appreciable levels in the sample that have close sequence similarity.
If you want to target known CDR3 sequences, please provide a FASTA file containing the CDR3. You may wish to also include some flanking sequence from framework regions (FWR3/FWR4) to ensure an optimal probe is picked.
For example, this representative PBMC dataset contains assembled CDR3 sequences that range from 42 bp to 48 bp. Here is an example assembled clonotype:
| Type | Sequence |
|---|---|
| Barcode | TATCAGGCACGCGAAA-1 |
| FWR3 | ACTGACCAAGGAGAAGTCCCCAATGGCTACAATGTCTCCAGATCAACCACAGAGGATTTCCCGCTCAGGCTGCTGTCGGCTGCTCCCTCCCAGACATCTGTGTACTTC |
| CDR3 | TGTGCCAGCAGCCGGGACAGGGTAAATCAGCCCCAGCATTTT |
| FWR4 | GGTGATGGGACTCGACTCTCCATCCTAG |
Converting this to a FASTA file, the inputs would look like this using 20 bp of each framework region:
>Clonotype1
CCCAGACATCTGTGTACTTCTGTGCCAGCAGCCGGGACAGGGTAAATCAGCCCCAGCATTTTGGTGATGGGACTCGACTCTC
Xenium In Situ chemistry can detect ssDNA or ssRNA at sufficient copy number. We recommend at least 2-4 copies per cell, but ideally ≥ 10 copies. The ideal number of copies per cell will depend on the proportion of cells expressing your barcode, the efficiency of the probe, and the importance that every cell contains the barcode you would like to detect.
A common use case for barcode detection is lineage tracing. Some use cases involve multiple barcodes in a combinatorial fashion. This is possible to read out with Xenium In Situ, however the probability that all of the events happen in a given cell decreases as the number of individual events that must be detected in a single cell increases.
Placing multiple copies of the same barcode on your construct will increase sensitivity by boosting the effective copy number. If you want to design probes in this fashion, it is ideal to put 50 bp of space between each barcode copy.