The Peak Viewer panel provides the tools to look at differential chromatin accessibility across genomic regions of interest. It combines two visualizations. The peak distribution chart shows the sizes of called peaks, and the relative number of cells per cluster within those peaks. The Peak Viewer also displays cut site tracks, which uses the fragments.tar.gz file generated by the Cell Ranger ATAC pipeline to show a finer-grained picture of chromatin accessibility on a base-by-base level. The Peak Viewer is exclusive to ATAC data and can be activated by clicking on the  icon to the left of the bottom data panel.
icon to the left of the bottom data panel.
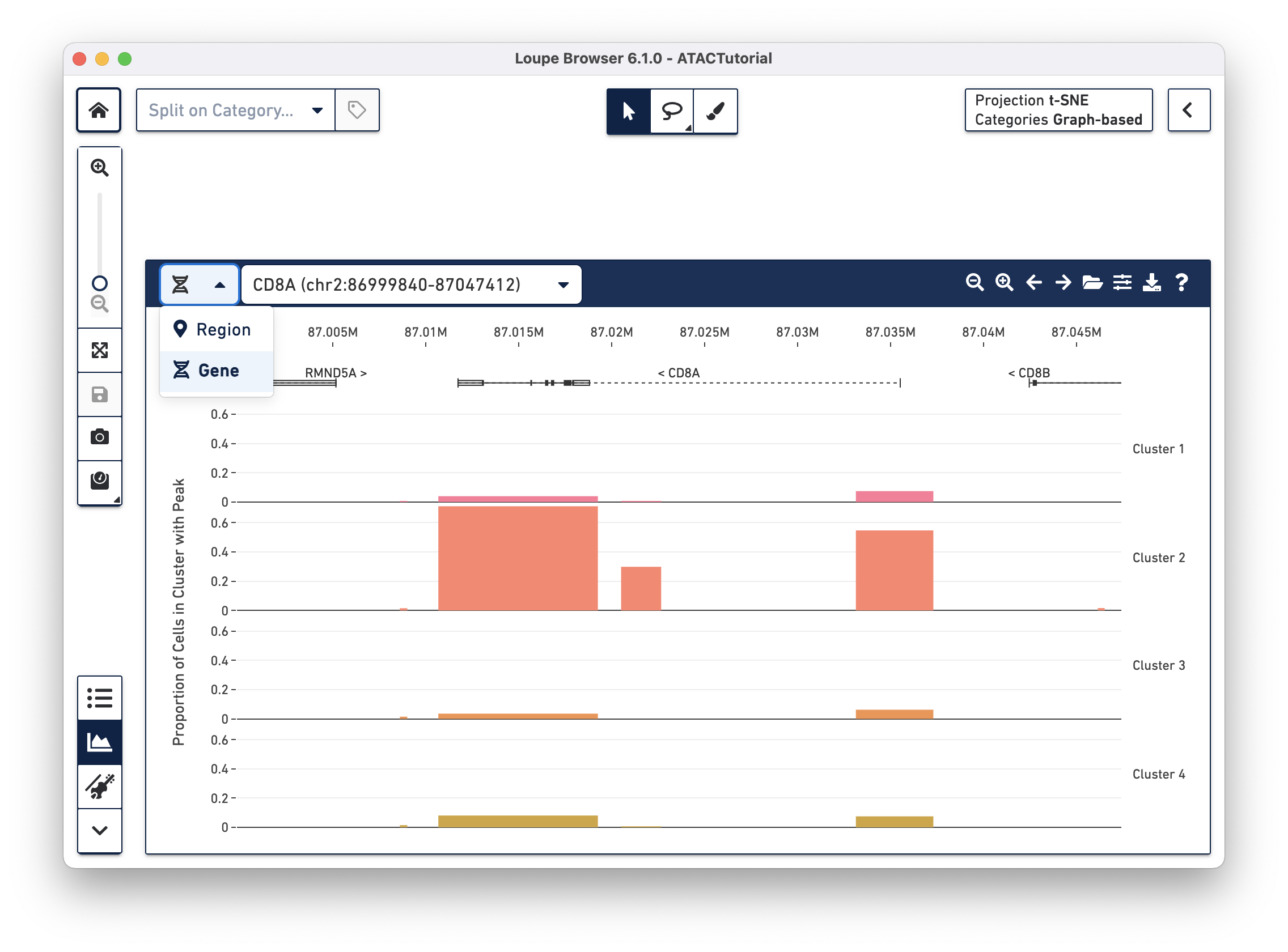
When loading a file for the first time, the Peak Viewer shows the peak distribution chart, with the window showing the first five called peaks in the dataset. You can see the currently selected range in the top left of the Peak Viewer ticks at the top of the panel to show genomic position, and a track of peak distributions per cluster. Called peaks appear as rectangles of varying width and height. The height of each peak is proportional to the number of cells within that cluster that had cut sites falling within the called peak. You can hover over each peak to identify the location of the peak, and the number of cells per cluster with a fragment within this peak. Clicking on a peak highlights the cells in the barcode plot where that region was accessible.
While peak distribution information comes preloaded in a .cloupe file, information about individual fragments, which drive the cut site tracks, are stored in another file generated by the Cell Ranger ATAC pipeline, the fragments.tsv.gz file. This file is bundled separately from the .cloupe file, as it usually takes up much more disk space. You can add the fragments file to the working session by clicking the folder icon on the Peak Viewer menu bar.
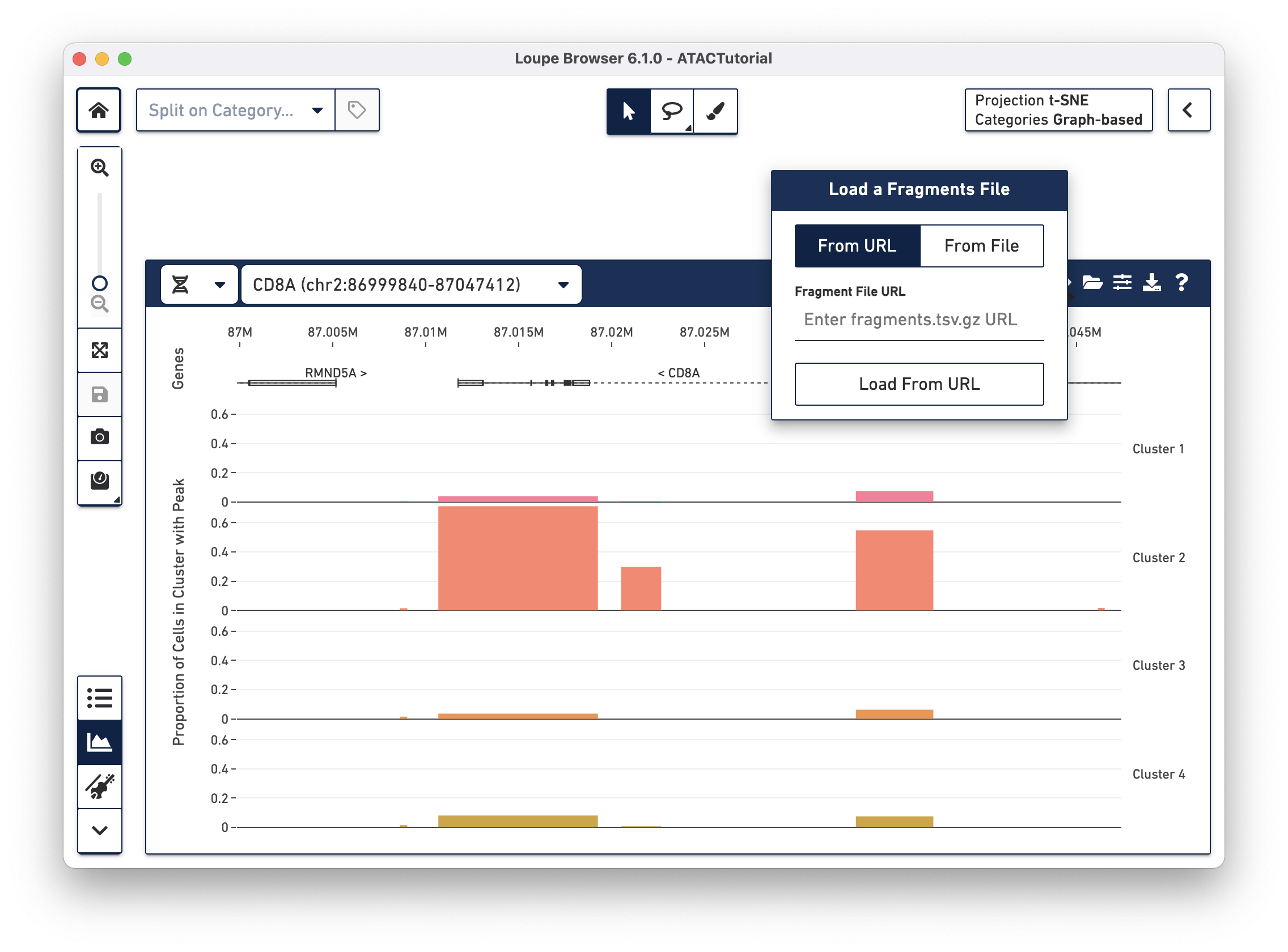
There are two options for attaching fragments files. If Cell Ranger ATAC outputs are available from behind a web server, you can specify the URL when the From URL tab is highlighted. For example, the URL of the ATACTutorial dataset's fragments.tsv.gz file is https://cf.10xgenomics.com/supp/cell-atac/ATACTutorial-fragments.tsv.gz.
Choosing the From File tab and clicking the Load From File button brings up your operating system's file chooser, through which you can navigate to select the appropriate fragments.tsv.gz file. If you provide an invalid URL, or the fragments.tsv.gz file selected is from a different dataset, you see a simple error message. If the association is successful, you see cut site tracks atop the peak distribution, and the peak distribution bars turn gray.
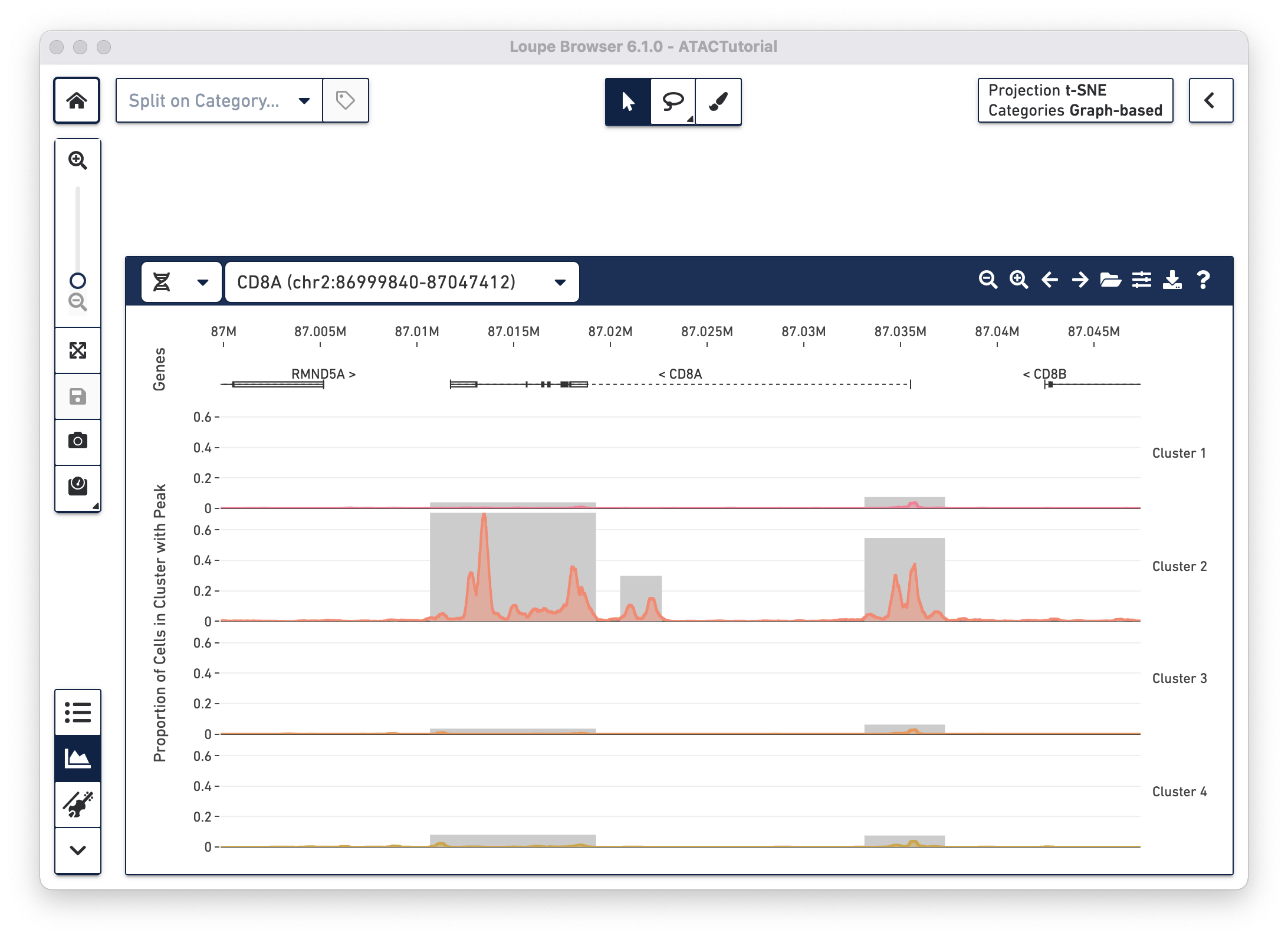
Shortcut: If you are loading the .cloupe file directly from a Cell Ranger ATAC outs folder, then Loupe Browser automatically detects the corresponding fragments file. If this is the case, you see both the peak distribution chart and cut site tracks in the Peak Viewer when first loading a file.
With the fragments file loaded, you can look at chromatin accessibility at base-level resolution by analyzing cut site tracks. Cut site tracks are generated by analyzing all of the fragments detected within the current viewing window from the fragments.tsv.gz file. Each read fragment called and validated by the Cell Ranger ATAC pipeline has two cut sites: the start position of the fragment, and the end position of the fragment. These are the locations where the ATAC enzyme is attached to open chromatin. For each cut site, the Peak Viewer imputes an open region, where the chromatin is presumed to be accessible. By default, the open window is 400 bases wide and centered around the cut site, though this is configurable (see Peak Viewer Options below). Finally, the cut site track is generated by computing a rolling sum over these open windows, akin to read pileups. In this manner, regions with the highest accessibility should have the highest number of nearby cut sites, the highest number of nearby open windows, and thus the highest rolling sum within the cut site track.
By default, the heights of the cut site tracks are normalized by the number of cells in each cluster, to highlight differences in accessibility between individual clusters. In this manner, the cut site tracks are measures of the average accessibility of a particular base for each cell in the cluster. You can also change this setting in the Peak Viewer Options menu.
With the cut site tracks visible, it is possible to associate the location of peaks with both genes and functional elements within genes. The Peak Viewer shows gene, transcript, exon, and UTR annotations in a bar above the cluster tracks. These annotations come directly from the reference specified in the Cell Ranger ATAC run. Genes are displayed as dashed lines with vertical line boundaries, the selected transcript as a solid line, exons as solid rectangles, and UTRs as open rectangles. In narrow window sizes, gene names are displayed above the annotation, though more information about a gene is available at any point by hovering over the annotation symbol.
Though there can be multiple transcripts per gene, the Peak Viewer only displays exons and UTRs from one transcript per gene. This is a visual compromise; there is limited screen space to show all transcripts from a reference. The heuristic for picking which transcript to display is as follows:
- A transcript must have one of the
appris_principaltags in the reference. - If multiple transcripts per gene have the
appris_principaltags, pick the ones that have the besttranscript_support_levelvalues (1 is best, 5 is worst). - If there are multiple transcripts with the same support level, pick the transcript sourced from HAVANA, rather than ENSEMBL.
- If there are still multiple HAVANA transcripts with the same support, pick the longest transcript.
- If there are still multiple well-supported primary HAVANA transcripts with equal length, then pick the first transcript by name as a tiebreaker (e.g., pick
TRANSCRIPT-002rather thanTRANSCRIPT-003)
Though only one transcript per gene is shown in the Peak Viewer, peaks can be marked as promoting genes if they are close to the transcription start sites of alternate transcripts. This is apparent by looking at CD8A below:
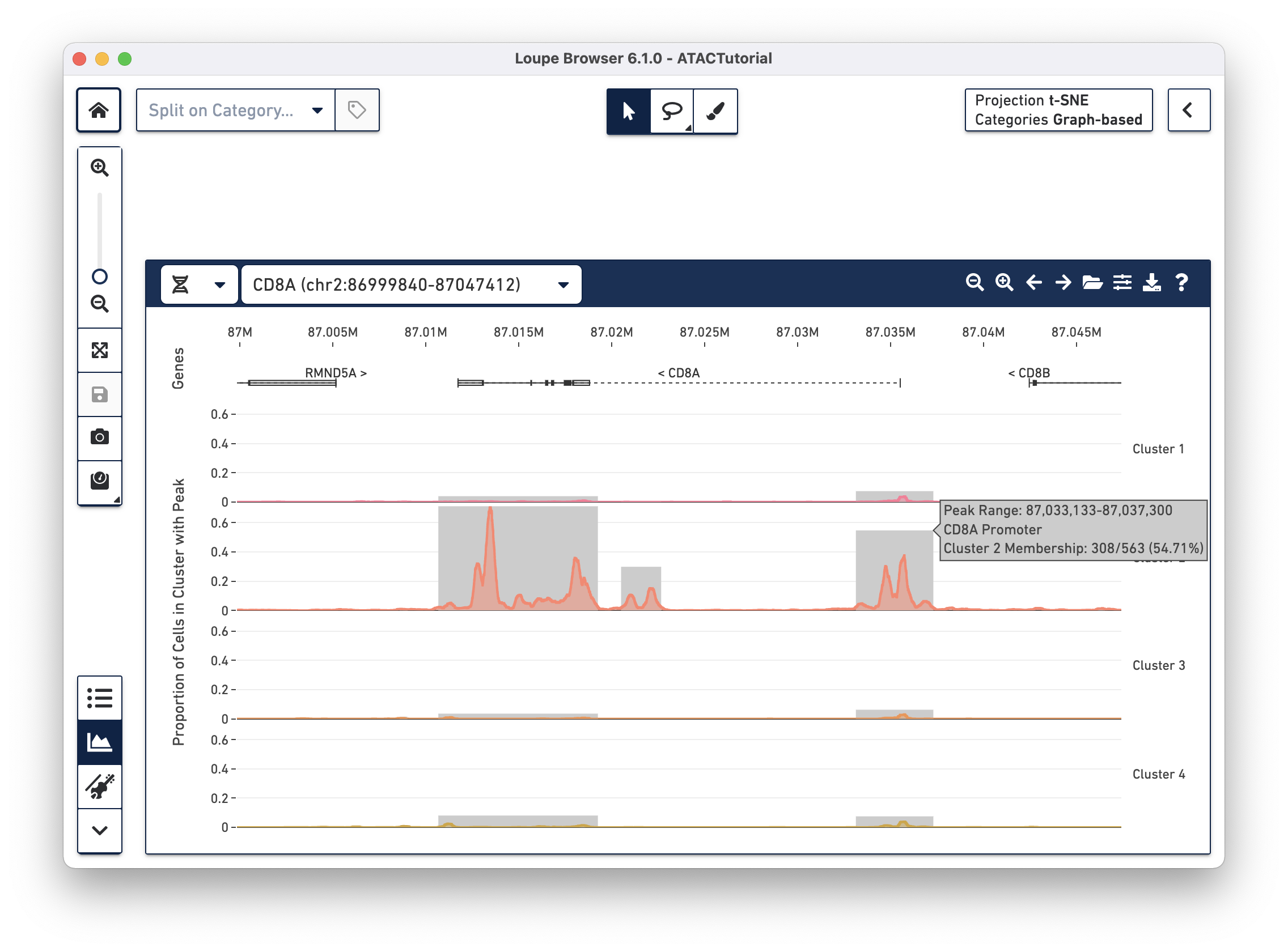
The rightmost peak within CD8A is close to the transcription start site of an alternate CD8A transcript specified in the reference, but not drawn in the annotation track.
Clicking on the sliders icon next to the folder icon brings up the Peak Viewer Options menu. The options include:
- Expand/Fit Peak Plots to Widow - Choose whether to expand the peak plot by hiding the genes and their coordinates at the top of the Peak Viewer.
- Hide/Show Peak Distribution - Choose whether to show the called peaks, either in colors (when cut site tracks are not visible) or in gray (when cut site tracks are visible).
- Hide/Show Cut Sites - With the fragments file loaded, choose whether to hide or show the cut site tracks.
- Disable/Enable Cut Site Normalization - By default, the cut site sums are normalized by cluster cell count to show the relative accessibility of a region between clusters (that may be of different sizes). Alternatively, you can see raw nearby cut site counts by choosing Disable Cut Site Normalization. The units displayed when hovering over the cut site track change accordingly.
- Show/Hide Annotations - Hide or show the gene and transcript annotations.
- Smoothing Window Size - Define the width of an open region around a cut site presumed to be accessible. Smaller values may change the apparent shape of a peak, and show more detailed accessibility patterns. You can show just the cut sites themselves by selecting a smoothing window size of 1.
You can export the peak distributions and cut site tracks from the Peak Viewer, for display or consumption by other bioinformatics tools. Click on the Export arrow icon to bring up the list of export options:
- Peak Distribution as CSV - Export the list of called peaks currently viewable in the window, and the number of cells per cluster with fragments in that peak.
- Cut Sites as BedGraph - Export the cut site tracks as a multi-tracked
.bedgraphfile. This can be viewed alongside other annotations in the UCSC Genome Browser. IGV does not support multi-tracked BED files; only the last cluster will be visible upon import. - Screenshot - Export the current state of the Peak Viewer window into either a PNG or SVG. It is recommended that you disable auto-height before export, for a cleaner, more expansive look.