Resources:
Fix, batch, and run later
Take advantage of robust fixation protocols that let you run assays on your schedule, simplify sample transport, and optimize your workflow.
Profile fresh, frozen, and fixed samples
Easily analyze previously inaccessible samples, from fixed tissues (even FFPE) to cells, nuclei, and FACS-sorted cells.
Reduce per-sample costs
Batch and multiplex to reduce per-sample costs by up to 75%, decrease sequencing cost, and maximize your resources.
Ultimate experimental flexibility
Process up to 1 million cells per run, change batch and sample throughput with ease, and capture protein alongside gene expression.
Industry-leading performance
Be confident in your data, using unmatched sensitivity for whole transcriptome profiling to analyze even damaged RNA or low-expressing genes.
Streamlined workflow and analysis
Explore gene expression profiles with reproducible and easy protocols, simplified data management, and rapid processing.
What you can do
Simplify sample logistics and set your own schedule
Sample fixation and storage allow for optimized and flexible sample batching while reducing variability and maintaining data quality.
Obtain high-quality data from challenging samples, even FFPE
Innovative probe design enables sensitive, robust performance, even from whole tissue, FFPE samples, and samples with fragmented RNA.
Bring cost-effective scRNA-seq to more samples
Increase the scope of single cell studies with reduced per-sample costs that make larger-scale experiments accessible.
Conduct multi-site experiments with ease
A reliable protocol preserves the biology of your precious samples, enabling research on a global scale.
Capture the data that fits your research needs
Assay flexibility lets you capture protein alongside gene expression, or target specific RNAs with user-designed probe pairs to supplement the whole transcriptome panel.
Proven Results
Videos
Resources
Robust results for scRNA-seq from matched FFPE and freshly fixed samples
Robust results for scRNA-seq from matched FFPE and freshly fixed samples
Data Spotlight
Data Spotlight
Isolation of Cells from FFPE Tissue Sections for Chromium Fixed RNA Profiling
Isolation of Cells from FFPE Tissue Sections for Chromium Fixed RNA Profiling
Demonstrated Protocol
Demonstrated Protocol
Single Cell Gene Expression Flex: Lock in cell states, unlock potential
Single Cell Gene Expression Flex: Lock in cell states, unlock potential
Blog
Blog
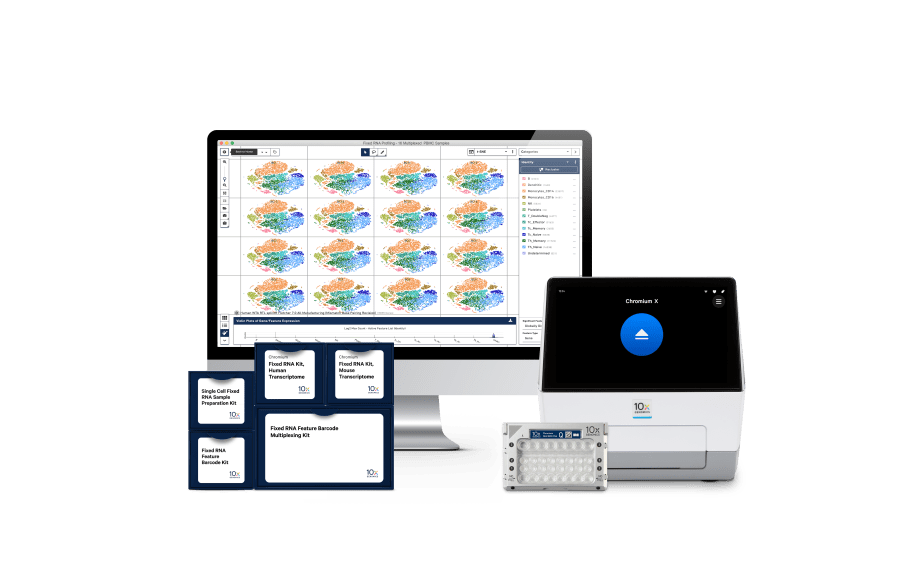
Our end-to-end solution

Success, right out of the box
The Chromium X handles all single cell assays, low to high throughput.

Chromium Single Cell Gene Expression Flex kits
With our reagent kits, explore cellular diversity through single cell, whole transcriptome analysis of fresh, frozen, or fixed samples for human or mouse, including simultaneous measurement of cell surface proteins.
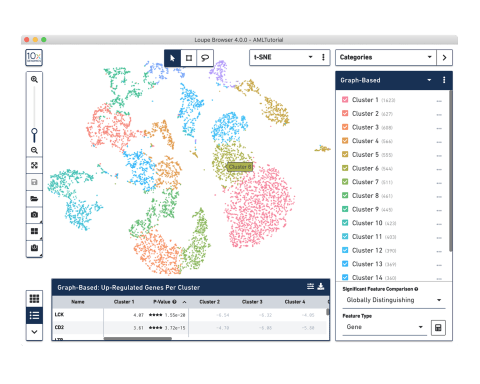
Intuitive analysis and visualization software
Discover single cell analysis, including cloud data processing, Cell Ranger analysis pipelines, and Loupe, our visualization app.

World-class technical and customer support
Our expert support team can be contacted by phone or email.
Workflow
Scroll to see the base workflow for Single Cell Gene Expression Flex. Options are included for sample multiplexing to increase throughput, or for use of Feature Barcode technology for cell surface protein detection in singleplex or multiplex workflows.
- 1
Collect your samples
Start with fresh, frozen, or fixed cells, nuclei, or whole tissue. For fresh samples, follow best practices for sample preparation to obtain a high-quality single cell suspension (or skip to Step 2 to fix tissue). If desired, stain whole cells with Feature Barcode technology to profile cell surface proteins simultaneously with gene expression.
For FFPE samples, follow our Demonstrated Protocol for guidance on how to dissociate FFPE tissue for use with this assay. After dissociation, samples can be safely stored at 4°C for up to 1 week or –80°C for up to 6 months. If not storing dissociated samples, you can instead move directly to Step 3.
Resources - 2

Fix and permeabilize samples
Follow our straightforward protocol to fix samples in paraformaldehyde (PFA), locking in biological states and preserving fragile cells. At this point, samples can be safely transported or stored at –80°C until you’re ready to work with them.
If working with FFPE samples, skip this section and move directly to Step 3.
Resources - 3
Construct your 10x library
Hybridize the probe set to your fixed samples, then partition individual cells on the Chromium iX or X instrument. After ligation and extension reactions, construct a 10x barcoded library. Within each nanoliter-scale partition, cells undergo RNA-templated ligation such that each resulting product shares a 10x Barcode from its individual cell of origin. For multiplexing workflows to increase throughput, hybridize your samples to probe sets, pool samples together, and proceed as usual—with up to 16 samples in a single lane of a Chromium chip. In this case, another unique barcode attached to each set of probes allows you to assign each individual molecule back to the sample it came from.
If, prior to sample fixation, whole cells were stained with Feature Barcode technology to profile cell surface proteins, a Protein Expression Library can be generated alongside the Gene Expression Library.
Resources- Chromium Fixed RNA Profiling – Protocol Planner
- User Guide for Singleplex Samples
- User Guide for Singleplex Samples with Feature Barcode technology for Protein
- User Guide for Singleplex Samples with Feature Barcode technology for Protein using Barcode Oligo Capture
- User Guide for Multiplexed Samples
- User Guide for Multiplexed Samples with Feature Barcode technology for Protein using Barcode Oligo Capture
- 4
Sequence
Resulting 10x barcoded libraries are compatible with standard NGS short-read sequencing on Illumina sequencers for massive transcriptional profiling of hundreds of thousands of individual cells at once.
Resources - 5
Analyze and visualize your data
Use our Cell Ranger analysis software to generate expression profiles for each cell, identify clusters of cells with similar profiles, and aggregate data from multiple samples. Interactively explore the results with our Loupe Browser visualization software.
Analysis pipelines output
Output includes QC information and files that can be easily used for further analysis in our Loupe Browser visualization software, or third-party R or Python tools.
Resources
Frequently asked questions
It enables the comprehensive profiling of the transcriptome (human or mouse) from single cells using a probe-based approach. The addition of Feature Barcode technology enables simultaneous profiling of cell surface protein expression for multidimensional insights into complex biology. This assay also enables sample multiplexing to increase your throughput and workflow efficiency.
In a direct comparison study, the Single Cell Gene Expression Flex assay was shown to be twice as sensitive as the Single Cell 3’ Gene Expression assay.
Fixation of samples preserves cell states, removing time constraints that can make single cell analysis challenging. The fixation protocol enables the transport of precious and/or fragile samples from the point of collection to central sites, such as core labs or service providers, for processing.
Fixation also enables greater efficiency in sample batching, which allows for more convenient workflows and reduces variability. Fixation can also neutralize infectious agents, and following proper validation of this step, samples may no longer need to be handled in a BSL-3 lab.
Human and mouse samples are compatible with this assay. Start with a fresh or frozen single cell/nuclei suspension or tissue and follow our fixation protocol, or start with FFPE tissue and follow our Demonstrated Protocol for guidance on dissociating FFPE samples for use with Single Cell Gene Expression Flex.
Fresh human and mouse samples are compatible with this option. Prior to fixation, whole cells can be labeled with antibody-oligonucleotide conjugates (using Feature Barcode technology) to profile cell surface proteins alongside gene expression.
10x Genomics Cloud Analysis enables you to process your single cell gene expression data through a simple web interface, leveraging an optimized, scalable infrastructure for fast results. Users can set up and run Cell Ranger pipelines through Cloud Analysis. Cell Ranger is a set of analysis pipelines that will automatically generate expression profiles for each cell and identify clusters of cells with similar expression profiles. Subsequently, Loupe Browser can be used to interactively explore the results.