The neXt generation of single cell RNA-seq: An introduction to GEM-X technology
Today, single cell RNA sequencing (scRNA-seq) is a well-established, reliable method enabling researchers to delve into the biological complexities driven by cellular heterogeneity. Scientists using scRNA-seq have made incredible discoveries, including identifying a neuronal population that helps paralyzed patients walk again (1); investigating cell–cell interactions driving implantation (the black box of human development) (2); and uncovering the mechanisms of action of an arsenal of therapeutics, including small molecules, immunotherapies, and cell therapies (3).
scRNA-seq is a technology driven by the need for cellular explorers to dive deeper into the complexities driving biological processes. The advent of scRNA-seq in 2009 (4)—like the launch of the NASA Hubble Space Telescope in the 1990s—gave us unprecedented views of an underappreciated universe. Since then, scientists studying everything from plant biology to drug toxicity have adopted scRNA-seq. But rapid and expansive adoption of emerging methods requires technological improvements to build new features and capabilities and accommodate increasingly complex research goals.
Eventually, space exploration required more discovery power than the Hubble offered, leading to the development of the James Webb telescope, which enabled scientists to see the birth of new stars at unprecedented resolution and more. Similarly, single cell exploration has outgrown the likes of the Hubble, and requires more powerful tools to keep moving research forward.
Our new, advanced GEM-X technology provides a robust foundation for expanding innovation and application support, ushering in the next generation of single cell. With optimizations to reagents and microfluidic chip architecture, the Chromium Single Cell Gene Expression (3’) and Chromium Single Cell Immune Profiling (5’) solutions are now more sensitive, more robust, and provide higher throughput, enabling more powerful insights in every facet of biology.
This new technology improves upon the fundamental chemistry fueling impactful single cell results. scRNA-seq adoption has become so widespread across fields such as developmental biology, cancer research, and neuroscience, even power users don’t always think about the reactions and microfluidics involved when performing pipetting steps that have become second nature, or using their instruments for the analysis of genes, cell by cell. (Perhaps they sometimes don’t remember what GEM stands for, like this blogger.)
In this blog, we’ll review the magic behind your UMAPs, the improvements GEM-X brings to these critical steps, and what those changes mean for your research. Read every chapter, or flip to the end:
- An overview of Gel Bead-in-emulsion (GEM) generation and barcoding
- The advantages of the end-to-end Chromium Single Cell workflow
- How GEM-X improves every scRNA-seq run
- What GEM-X means for your research
How does 10x Genomics scRNA-seq work?
The basic chemistry that drives the previous iteration of our Chromium Single Cell Gene Expression and Single Cell Immune Profiling assays, powered by Next GEM technology, remains the same in our GEM-X assays—it is related to the foundational Drop-seq method published by Harvard researchers in 2015 (5).
Like most great science, the process begins with a researcher pipetting microliters of seemingly identical clear liquids into a microfluidic chip with oil to form reaction vessels called Gel Beads-in-emulsions (GEMs).
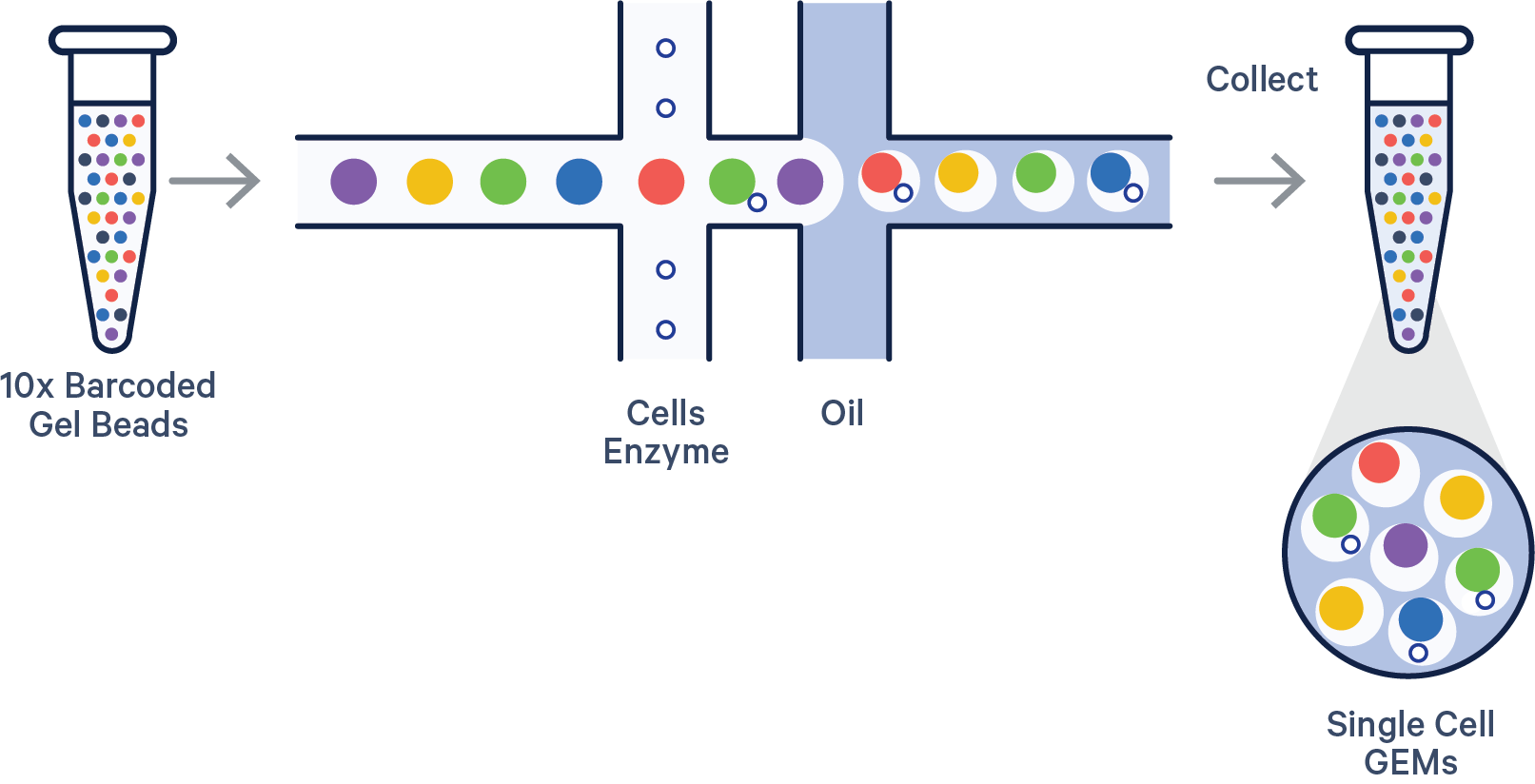
But these clear liquids are far from identical. The mixture is composed of a cell suspension with lysis buffer, Gel Beads coated with oligonucleotides—which include a 10x Barcode that marks each RNA molecule’s cell of origin and a unique molecular identifier (UMI) that gives each transcript a unique fingerprint—a poly(dT) sequence for capturing mRNA at the 3’ or 5’ end, and adapter sequences for downstream next-generation sequencing.

Once the chip is placed in our Chromium X Series instrument, cells move through a microfluidic labyrinth at a limiting dilution to generate GEMs, which appear as nanometer-sized rain drops, each containing a single cell. The cell is lysed and the Gel Bead dissolves. The 3’ end of the released RNA binds to the poly-dT sequences that coated the Gel Bead. The primed RNA is reverse transcribed, forming complementary DNA (cDNA).
All cDNAs from a single cell, originating from a single Gel Bead, will have the same 10x Barcode, enabling the user to map each transcript back to its cell of origin. The individual oligos on a single Gel Bead, however, have different UMIs, which distinctly mark each cDNA molecule with a unique, random 12-base sequence. This allows the user to eliminate any PCR duplicates during downstream processing.
The barcoded cDNA is then used to prepare a sequencing library, which includes the addition of sequencing adapters and PCR amplification. Finally, the library is sequenced with NGS.
Our Single Cell Immune Profiling assay utilizes primers targeting the 5’ end of mRNA, and works a little differently. We will delve into those differences in a future blog dedicated to the multiomic capabilities of this powerful single cell solution, now powered by GEM-X.
What are the benefits of conducting scRNA-seq analysis with the proven, instrument-supported Chromium workflow?
The Chromium platform standardizes the scRNA-seq workflow, making reliable, reproducible, and impactful single cell insights accessible to every researcher, no matter their skill level or expertise.
Scientists in countless research areas, from neuroscience to immunology to drug development, have used our Chromium Single Cell Gene Expression (3’) and Single Cell Immune Profiling (5’) assays to make incredible discoveries, as evidenced by over 6,500 published studies, including nearly 700 papers in Nature, Science, and Cell.
A key aspect of our technological innovation is our instruments. Our Chromium Single Cell assays are supported by instruments that automate the most crucial step in any scRNA-seq experiment—cell partitioning and barcoding. With advanced algorithms and bioinformatics tools, researchers can efficiently process sequencing data, transforming raw reads into actionable insights.
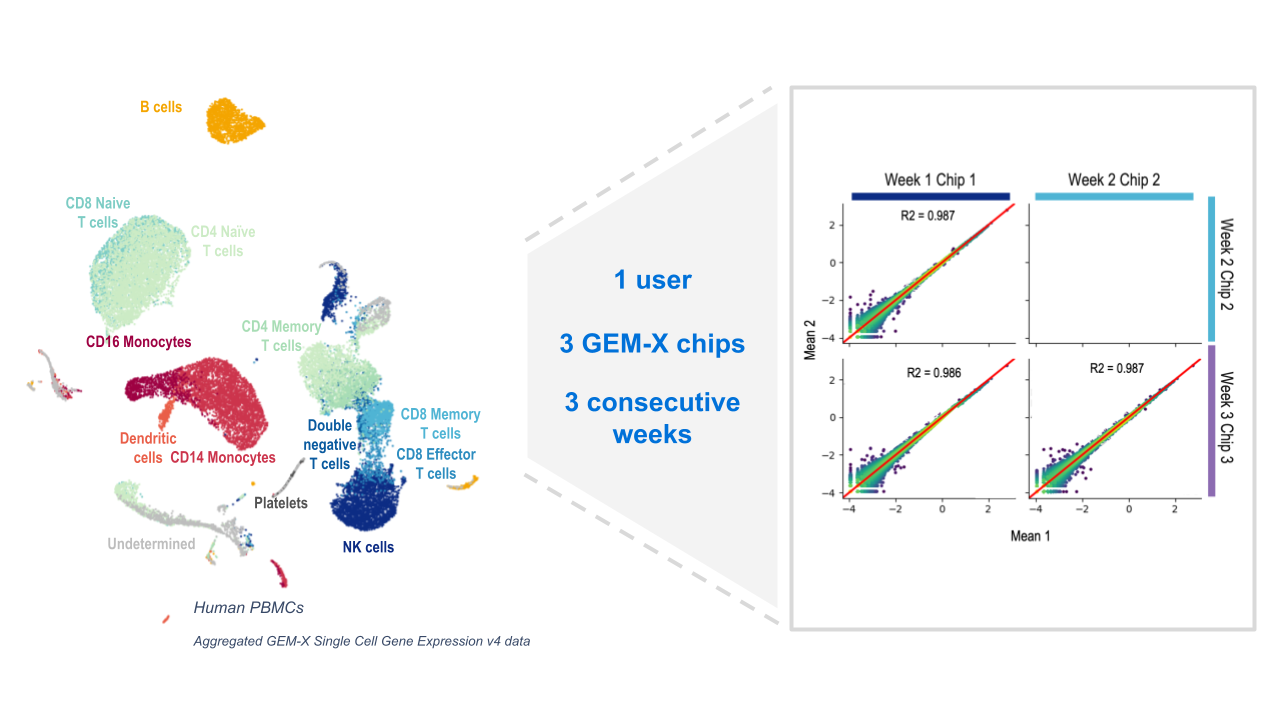
While all single cell workflows require instrumentation such as PCR machines and sequencers, automating the partitioning and barcoding step limits the potential for technical error or batch effects introduced by a manual workflow with lots of hands-on time and pipetting steps. Different researchers with different skill sets (even across multiple institutions) can conduct the same experiment using a robust platform that helps them get the same results.
How does GEM-X improve upon the tried-and-tested Chromium scRNA-seq workflow?
With GEM-X, we’ve improved every step of our tried-and-tested scRNA-seq workflow with optimized reagents, improved microfluidics, and upgraded analysis software supported by our most advanced single cell instruments—the Chromium X Series.
We redesigned the GEM-X microfluidic chip architecture, settings, pressure, partition size, and more to improve the overall performance of our Chromium Single Cell Gene Expression (3’) and Single Cell Immune Profiling (5’) assays compared to previous versions—including minimum clogging and maximized cell size flexibility. This redesign improved key steps in our single cell workflow, including GEM generation and cell partitioning.
While our Next GEM microfluidic chips require step emulsification—whereby GEMs are generated independently of partitioning oil—GEM-X microfluidic chips utilize the flow of partitioning oil to facilitate GEM formation. Twice as many GEMs are generated at smaller volumes, reducing multiplet rates two-fold and increasing throughput capabilities.

GEM-X microfluidic chips also enable faster and more robust cell partitioning. The reduced 6-minute run time has no negative impact on cell stress as the cells quickly maneuver through GEM-X chips, enabling retention of fragile cell types.
But what do these technical upgrades mean for your research? Higher performance across the board.
How can high-performance scRNA-seq, powered by GEM-X, bolster my research?
The advanced GEM-X technology powering our next generation of Chromium Single Cell Gene Expression (3’ v4) and Single Cell Immune Profiling (5’ v3) solutions is not only the culmination of our 10+ years of experience building microfluidic chips and developing industry-leading single cell assays—it’s also the direct result of customer feedback. This was a product built for our customers, with the goal of enabling them to take their research to new heights.
Before we dive into the data, let’s highlight the key advantages of GEM-X compared to Next GEM technology:
- Increased sensitivity enabled by a two-fold increase in detected genes and improved capture of rare transcripts, fragile cells, and cells with low RNA content
- Higher throughput with a two-fold increase in cells captured per channel (up to 20,000 cells per channel)
- More cost effective with a more than 50% reduction in cost per cell and sample
- Improved sample recovery—up to 80% cell recovery efficiency
- Enhanced data quality and robustness due, in part, to a halved multiplet rate (0.4% per 1,000 cells) and more efficient GEM generation with no increase in cell stress
- Faster run times—tens of thousands of cells are partitioned and barcoded in just 6 minutes
Why do improved gene sensitivity and cell recovery matter?
scRNA-seq enables researchers to unmask rare transcripts, cell states, and cell populations other methods, such as bulk RNA-seq, flow cytometry, and mass spectrometry, can’t detect. With single cell resolution, researchers can discover novel biology they didn’t know to look for.
But capturing the critical insights buried in sample complexity requires methods that can both capture those rare cells and sensitively detect the lowly expressed transcripts they hold.
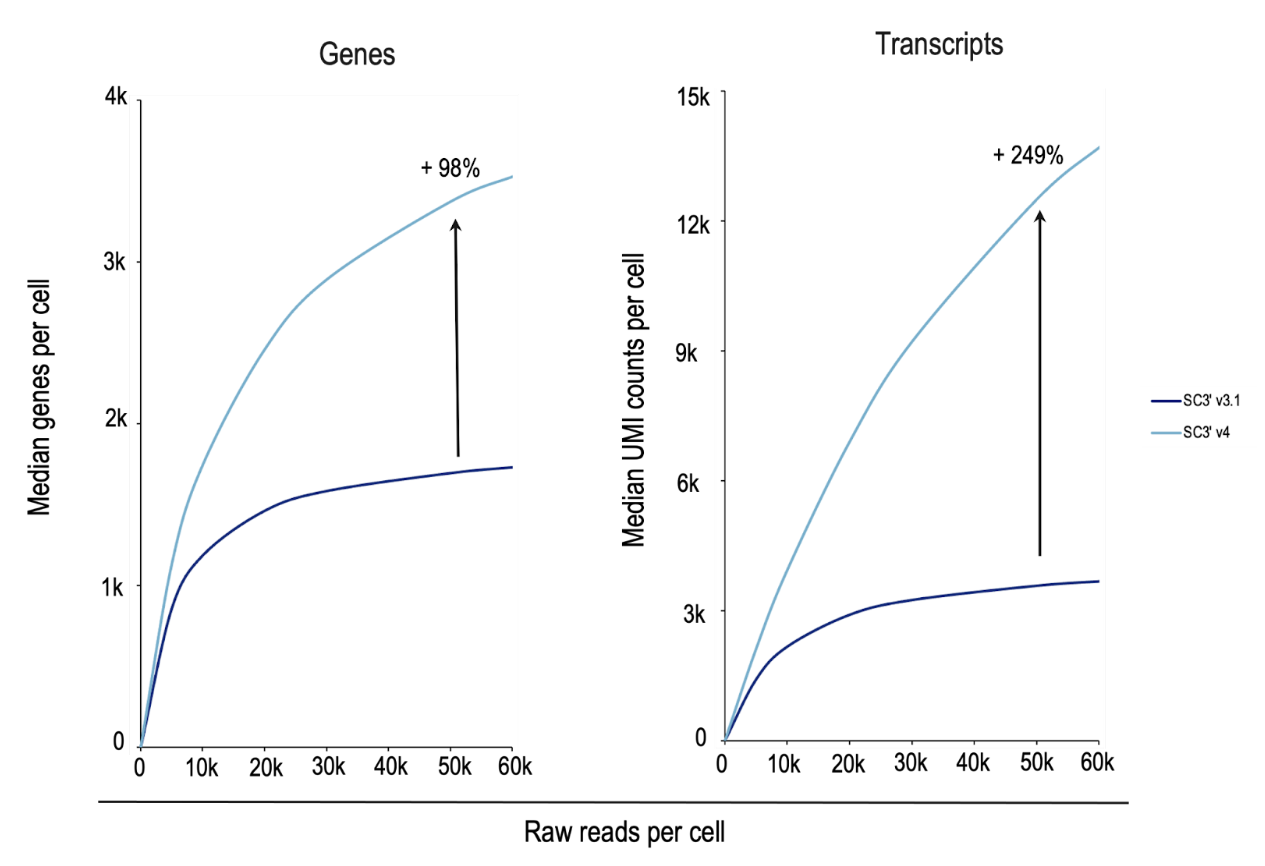
In a side-by-side experiment analyzing nuclei from complex mouse brain samples (Figure 6), Chromium GEM-X Single Cell Gene Expression v4 detected 98% more genes and 249% more transcripts per cell than Chromium Next GEM Single Cell Gene Expression v3.1. Chromium GEM-X Single Cell Immune Profiling v3 is also more sensitive than its Next GEM predecessor (v2)—side-by-side analysis of human peripheral blood mononuclear cells (PBMCs) with GEM-X detected 61% more genes and 103% more transcripts (data not shown).
What could this mean for your research? With increased sensitivity, researchers can detect cells in short-lived transient states, enabling critical insights during differentiation. When cells differentiate from one cell to another, genes are rapidly and dynamically regulated to move the cell forward to the next stable cell state—like the transition from a neural progenitor cell to a neuron. Cell surface markers don’t always change in a significant way between one cell type to another, leaving these transient states undetectable with commonly used techniques like flow cytometry.
With highly sensitive single nuclei or single cell RNA-seq, however, researchers can analyze transcript levels from individual cells in samples with actively differentiating cells, giving researchers the resolution needed to parse transient cell states. Identifying these short-lived transitions can help improve our understanding of developmental biology, aging, and identify the most efficacious cell populations for cell therapies.
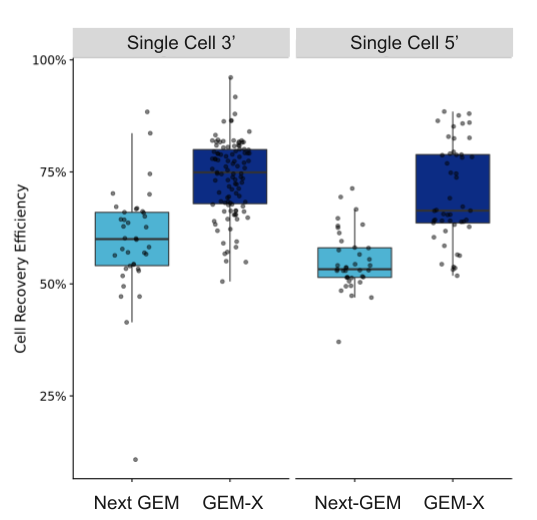
Researchers also have a better chance at identifying these critical, but sparse, cell states if they capture more cells from their samples. As shown in Figure 7, both our Chromium GEM-X Single Cell Gene Expression v4 (3’) and Chromium GEM-X Single Cell Immune Profiling v3 (5’) assays have improved cell recovery efficiency—up to 80% with GEM-X Single Cell Gene Expression v4 compared to 65% with Next GEM Single Cell Gene Expression v3.1—not only enabling improved detection of rare cell populations, but better capture of cells in precious samples, including samples typically yielding few cells like tissue biopsies or previously flow sorted cells.
For example, single cell technology critically helps scientists break down the complexity of the tumor microenvironment—delineating mechanisms driving metastasis, drug efficacy, and side effects—through analysis of patient tumor samples. More powerful single cell tools offer enhanced discovery of rare cell populations in heterogeneous tumor samples from patients, enabling identification of mechanisms of drug efficacy, therapeutic resistance, and toxicity, ultimately informing more effective patient stratification.
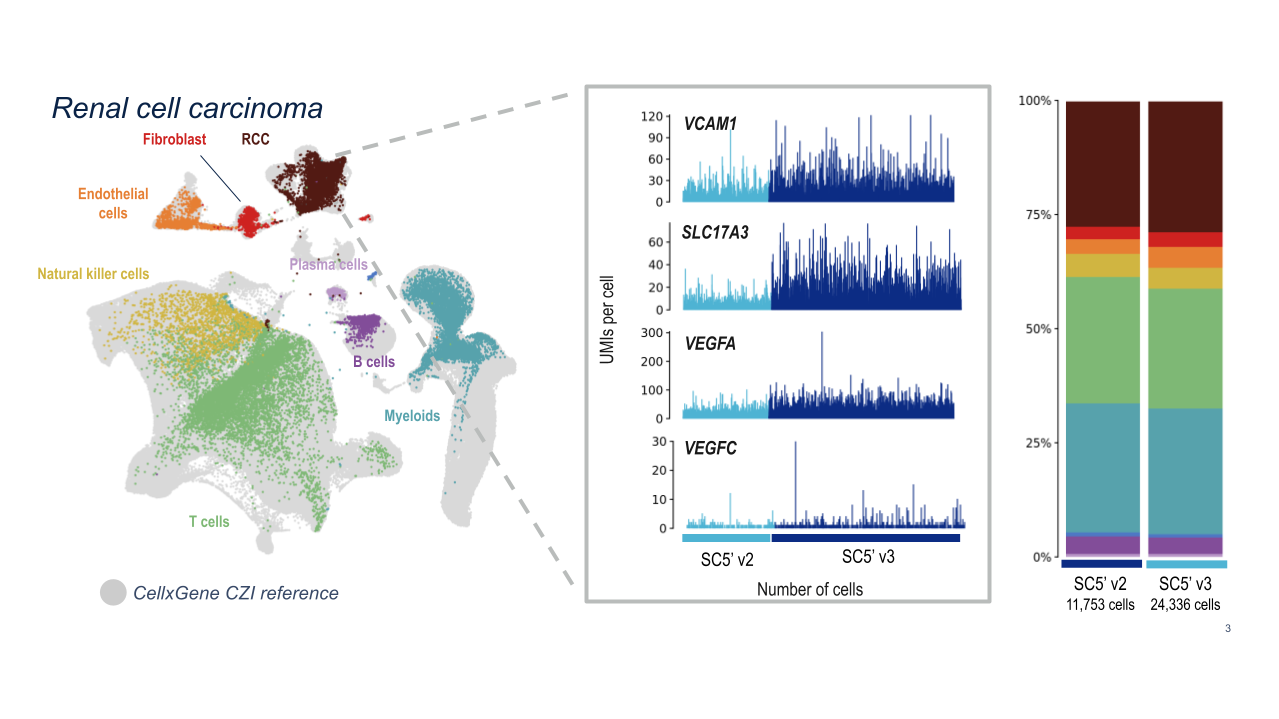
10x researchers demonstrated the powerful analytical capabilities of Chromium GEM-X Single Cell Immune Profiling (5’) v3 for clinical samples by analyzing dissociated tumor cells from human renal cell carcinoma samples. GEM-X consistently detected four key markers of renal cell carcinoma at higher levels than Chromium Next GEM Single Cell Immune Profiling (5’) v2.
How can cost-effective scRNA-seq solutions with higher throughput boost your study’s impact?
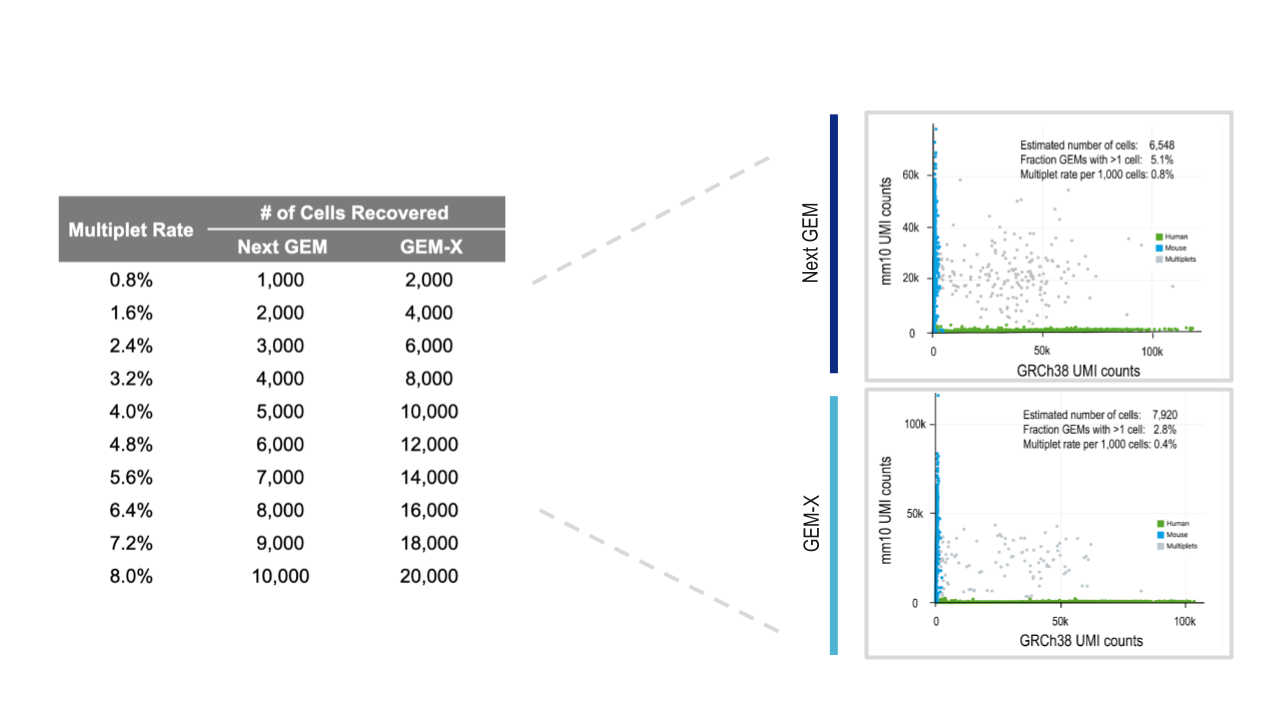
Our Chromium GEM-X Single Cell Gene Expression (3’) v4 and Chromium GEM-X Single Cell Immune Profiling (5’) v3 assays further conserve your precious samples by enabling higher-throughput and more cost-effective analyses. You can now run 20,000 cells per lane at more than half the cost per cell, with a 2-fold lower rate of multiplet formation compared to Next GEM–powered assays, Chromium GEM-X Single Cell Gene Expression (3’) v3.1 and Chromium GEM-X Single Cell Immune Profiling (5’) v2.
Improved cell capture rate, low multiplet formation rates, and higher throughput capabilities increase the likelihood of capturing rare cell populations critical for basic processes or therapeutic efficacy and resistance. In fact, a 2020 survey of single cell transcriptomic publications demonstrated that the number of cell types identified correlated strongly with the number of cells studied (6).
Scaling up your studies enables more than highly robust analysis, however. Including more samples can increase the statistical power of your study, making it both more insightful and reliable.
But scaling up studies to hundreds of thousands to millions of cells can be cost-prohibitive.
That’s why we are committed to making single cell analysis more accessible by reducing the cost per cell and sample. Many use single cell to validate their bulk RNA-sequencing results because of resource limitations. But with higher-throughput, cost-effective scRNA-seq, researchers don’t have to limit their studies or their scientific impact. And they gain a large dataset to mine or return to in the future to answer new questions or drive other scientific inquiries.
What cells have you been missing? How does GEM-X help you detect fragile, low RNA–content cells?
GEM-X expands discovery potential with increased detection of fragile, low RNA–content cells. Because cells move more quickly through the channels in GEM-X microfluidic chips, fragile cells spend less time in suspension and are preserved; high sensitivity ensures cells with low RNA–content are detected.
Capturing these cells increases your chance of finding the cell you didn’t know you were looking for—or the cell you knew you were missing.
Neutrophils are notoriously challenging to capture for scRNA-seq analysis due to their fragile nature, low RNA content, and, potentially, the presence of interfering proteases and nucleases in neutrophil granules—a factor that makes all granulocytes difficult to capture (7). We here at 10x and others have developed modifications to existing scRNA-seq protocols and data analysis strategies to increase the number of detected neutrophils, but capturing these finicky cells remained technically challenging.
Chromium GEM-X Single Cell Gene Expression (v4) removes this technical challenge, and is able to efficiently capture neutrophils with the standard workflow. When 10x researchers compared scRNA-seq analysis of fresh human leukocytes with Chromium GEM-X Single Cell Gene Expression (v4) against analysis with Chromium Single Cell Gene Expression (v3.1), they detected a substantially larger proportion of neutrophils.
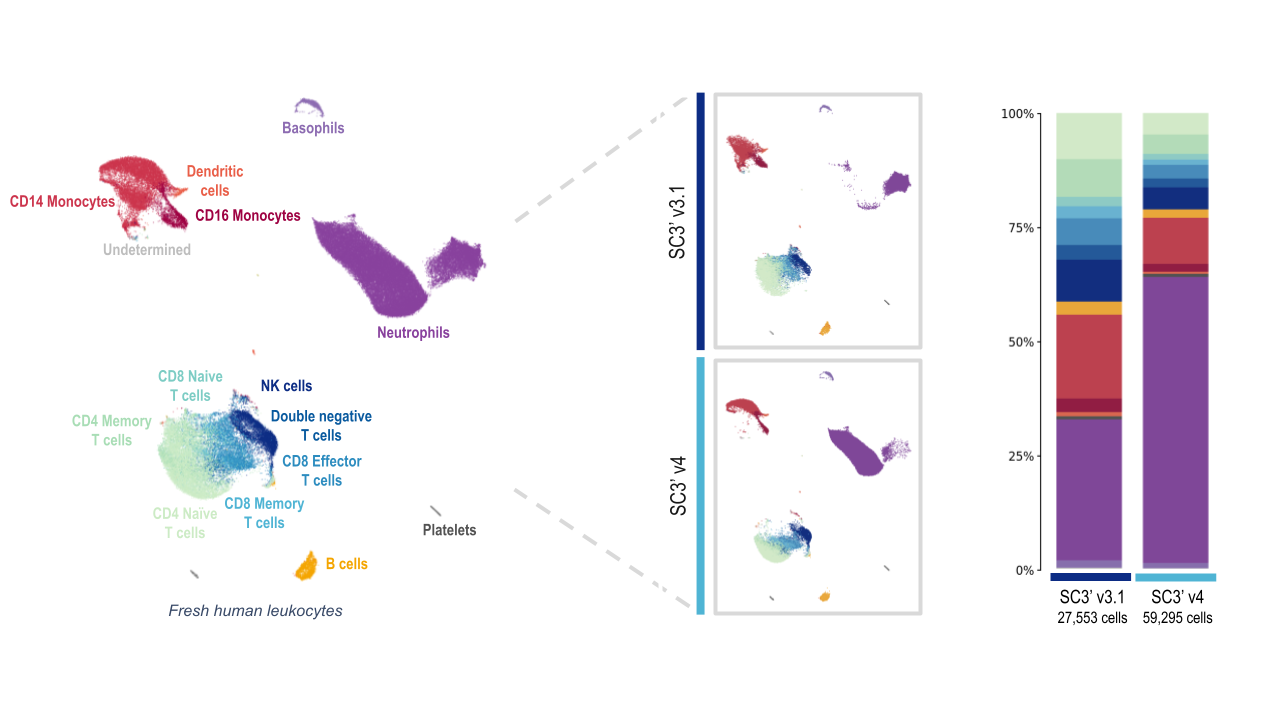
Both the Next GEM (3’ v3.1) and GEM-X (3’ v4) assays captured all the expected major cell-type populations, including several T-cell populations, B cells, and platelets. At first glance, the data from leukocyte analysis with the GEM-X Single Cell Gene Expression v4 assay looks skewed. But this is largely due to the massive increase in the amount of neutrophils detected.
Both chemistries captured a similar number of cells of each major immune cell type, with the exception of the neutrophil population. The Chromium GEM-X Single Cell Gene Expression v4 assay detected nearly quadruple the number of neutrophils as the Single Cell Gene Expression v3.1 assay, powered by Next GEM technology.
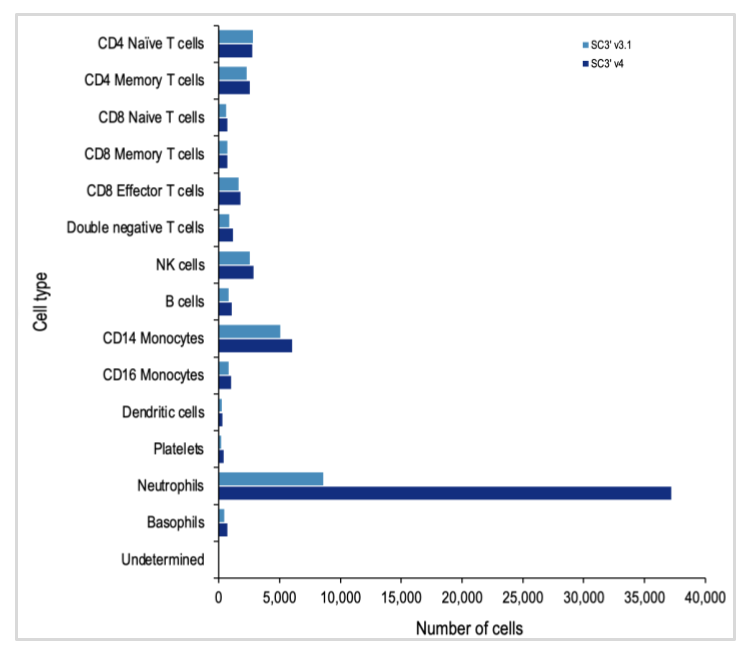
Since neutrophils are the most abundant cell type in the blood, this opens the door to exciting new discoveries related to every facet of the blood and immune responses, enabling new insights into immune responses to infections, vaccine development, and mechanisms and druggable pathways for autoimmune disorders.
How can immune profiling assays benefit from GEM-X performance improvements?
Capturing neutrophils isn’t the only way GEM-X will enhance immunological studies—improvements to our Chromium GEM-X Single Cell Immune Profiling assay will help researchers better understand the complex mechanisms driving immune functions.
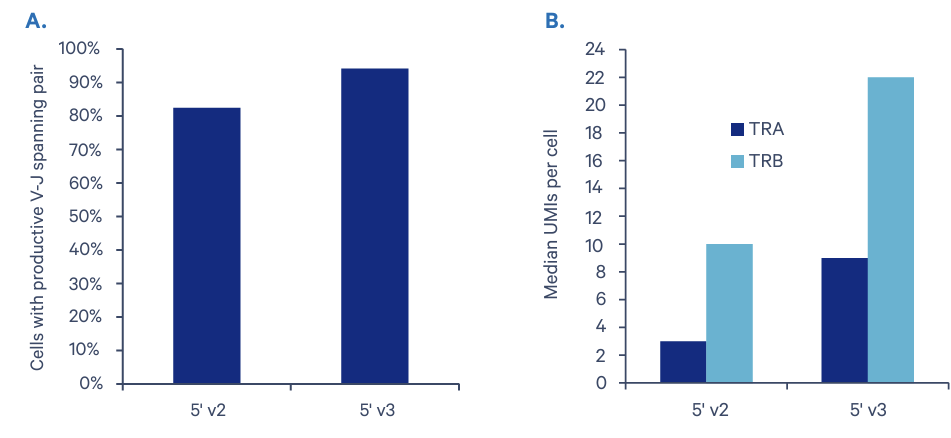
A key feature of this assay, comprehensive analysis of the immune cell repertoire with full-length V(D)J sequences for paired B-cell or T-cell receptors (BCRs or TCRs), is significantly improved with Chromium GEM-X Single Cell Immune Profiling (5’) v3. The average V-J pairing rate is 95% compared with the 84% rate obtained with our Chromium Single Cell Immune Profiling (5’) v2. Our research team also observed a 200% and 120% increase in detected expression of T-cell receptor genes TRA and TRB, respectively.
Enhanced characterization of BCRs and TCRs will help maximize insights into B- and T-cell function in health, disease, and drug and vaccine immune responses. This is particularly important as we improve our ability to handle future outbreaks of emerging diseases with lessons learned from the ongoing COVID-19 pandemic.
The neXt generation of single cell is just getting started
The technological advances discussed in this blog are just the beginning of the GEM-X era of single cell technology. GEM-X is a robust foundation for expanding innovation and application support on the Chromium platform, allowing you to plan for the future of your research. We’re excited to further discuss improvements to our Single Cell Immune Profiling assay and its other multiomic capabilities—including cell surface protein (also available with our 3’ assay) and CRISPR perturbation screening—in future GEM-X-focused blogs.
As we keep enhancing the capabilities of our assays and building out new features, you can learn more about our GEM-X-powered Chromium Single Cell Gene Expression and Single Cell Immune Profiling solutions by downloading our resource toolkit here.
References:
- Kathe C, et al. The neurons that restore walking after paralysis. Nature 611: 540–547 (2022). DOI: 10.1038/s41586-022-05385-7
- Xu Y, et al. A single-cell transcriptome atlas profiles early organogenesis in human embryos. Nat Cell Biol 25: 604–615 (2023). DOI: 10.1038/s41556-023-01108-w
- Crees ZD, et al. Motixafortide and G-CSF to mobilize hematopoietic stem cells for autologous transplantation in multiple myeloma: a randomized phase 3 trial. Nat Med 29: 869–879 (2023). DOI: 10.1038/s41591-023-02273-z
- Aldridge S & Teichmann SA. Single cell transcriptomics comes of age. Nat Commun 11: 4307 (2020). DOI: 10.1038/s41467-020-18158-5
- Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161: 1202–1214 (2015). DOI: 10.1016/j.cell.2015.05.002
- Svensson V, et al. A curated database reveals trends in single-cell transcriptomics. Database 2020: baaa073 (2020). DOI: 10.1093/database/baaa073
- Wigerblad G, et al. Single-cell analysis reveals the range of transcriptional states of circulating human neutrophils. J Immunol 209: 772–782 (2022). DOI: 10.4049/jimmunol.2200154
About the author:

