When single cell meets spatial: Novel lung immune niche provides long-term antibody hideouts
What happens when two powerful technologies join forces to resolve the “what” and the “where” of complex biology? We share those stories in our “When single cell meets spatial” series, highlighting studies that capture the best of both technologies—and how they can be even better together.
While single cell gene expression analysis provides a high-resolution view of the cellular heterogeneity of a complex sample, scientists lose key contextual information by digesting a sample into a single cell suspension. With the addition of spatial transcriptomics, researchers can not only access crucial information about cellular localization, cell–cell interactions, and cell signaling, but also reinform cell type–identification in morphological context with the resolution provided by single cell assays.
We’re still learning about lung immunity
From the annual cold to the COVID-19 pandemic, trends in infectious disease show us again and again that the lungs represent the immunological battleground of the body. And the stakes in the battle of virus versus host are high.
“Not only do the lungs represent the environment most frequently targeted by pathogens, their role as the organ of gas exchange makes their normal functioning critical for health and intolerant of collateral damage” (1).
Resident innate and adaptive immune cells, as well as specialized respiratory epithelial cells, give our lungs defensive firepower against infectious agents (1), but we’re still learning about the breadth of cells that perform these protective functions. In their recent Nature Genetics publication, researchers from the Wellcome Sanger Institute established a single cell and spatial atlas of healthy human lungs and airways, revealing a novel immune cell niche for IgA plasma cells that likely supports respiratory health (2). Their findings, available as a public dataset, include 80 distinguished cell types and states and 11 populations not previously annotated, representing a major contribution to the Human Lung Cell Atlas.
Keep reading to learn how combining single cell and spatial gene expression technology enabled these discoveries, and how they promise to bring a new dimension to our understanding of cellular heterogeneity and immunity in the lungs.
Mapping known, rare, and novel cells in the lungs and airways
The lung’s essential functions, including gas exchange and immunosurveillance, are ultimately maintained by cells—many diverse cell types and their coordinated relationships with one another. Together, these cells build the tissue architecture of the respiratory system.
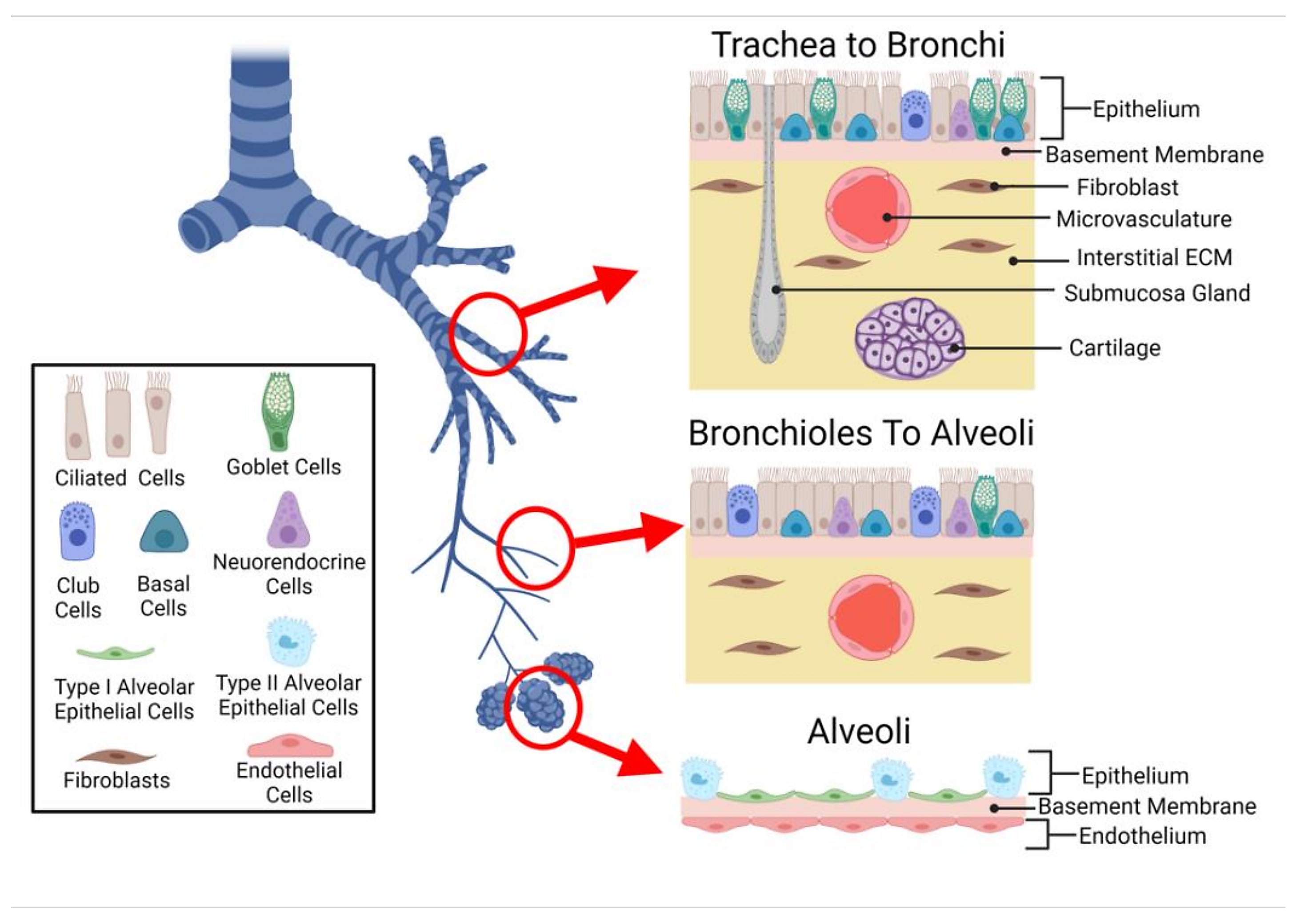
To explore the complex cellular microenvironments of the human lungs and airways, scientists from Wellcome Sanger Institute performed deep tissue profiling on healthy human trachea, bronchi, and upper and lower parenchyma samples using single cell/nucleus RNA-seq (sc/snRNA-seq) and Visium spatial transcriptomics. Their spatial analysis included 20 tissue sections. Using the cell2location algorithm, they used their single cell data to deconvolve, or inform, their Visium data—in their words, determining “which cell types from suspension data in which abundance could explain the mRNA counts observed in the Visium data (2).”
Their combined analytical approach enabled the team to not only characterize expected cell populations, but reveal rare cell types, too. They characterized an immune recruiting fibroblast population (IR-fibro) that localized with a rare immune infiltrate microenvironment in the bronchus. Using IR-fibro marker genes, which include chemokines responsible for T- and B-cell recruitment in secondary lymphoid organs, the team detected this cell type in existing lung cell atlas data. They described a rare and previously uncharacterized macrophage subtype distinguished by its unique chemokine expression profile, linking it to macrophage populations associated with lung infection, asthma, and psoriasis. The team also described, for the first time in human lungs, a CD11d+ natural killer cell population typically seen in the blood and activated in response to infection (2).
With this previously inaccessible resolution of immune cells in human lungs, the research team was able to trace a population of IgA antibody-secreting plasma cells to submucosal glands (SMG) in the trachea. There, the plasma cells localized with duct, mucous, and serous cells, which make up the SMG. Performing spatial transcriptomics analysis on formalin-fixed paraffin-embedded (FFPE) preserved tracheal samples, which provided a refined view of serous and mucous glands, and immunohistochemistry, they confirmed a pattern of preferential localization for these IgA plasma cells in the SMG. In contrast, an IgG subtype was excluded from the SMG and localized instead with immune infiltrates (2).
Durable antibody immunity—a key factor of lung health
Mucosal IgA is a known, important factor for protection against respiratory infections (2). “The development of resident antibody-secreting cells following respiratory virus infection or vaccination is key to the establishment of durable immune protection against airway pathogens,” said Sealy et al (3).
The research team confirmed the importance of IgA by looking back at existing single cell data from COVID-19 patient lung samples—there was a clear increase in IgA plasma cell abundance in COVID-19 patients compared to healthy controls. Given this evidence, the team concluded that IgA plasma cells, together with a cohort of naive B cells and CD4+ T helper cells characterized in the SMG, form a gland-associated immune niche that is likely highly relevant to disease (2).
But they took this finding one step further. Using cell–cell interaction analysis, enabled by spatial data and a tool called CellChat, the research team uncovered an axis of cellular communication between SMG epithelial cells and B plasma cells that predicted SMG duct cells serve to recruit memory and naive B cells and CD4 T cells through chemokine ligand 20 (CCL20). Additionally, SMG duct and serous cells were shown to express A proliferation-inducing ligand (APRIL), which, when interacting with surface receptors on B cells, supports B-cell survival, differentiation, and class switching (2).
These insights into the cellular and molecular pathways that generate and support immune recruitment and antibody secretion in the context of antiviral respiratory immunity provide the foundation to find answers to a number of subsequent questions:
- What are the fundamental contributions of these immune niches and their core functions in long-term antiviral protection?
- How do we best design vaccines to engender durable antibody secretion in these resident lung immune niches?
- What may go awry in these systems that could contribute to hyperinflammation, or possibly the cytokine storm and resulting tissue damage, or acute respiratory distress syndrome?
The resolution of these questions isn’t clear yet, but the immunology and infectious disease community is one step closer to understanding with a comprehensive single cell and spatial lung atlas in hand.
To explore this research further, review the publication here and navigate to the team’s dataset or the Human Lung Cell Atlas. And find resources for Chromium Single Cell Gene Expression or Visium Spatial Gene Expression.
References:
- The lungs at the frontlines of immunity. Nat Immunol 16: 17 (2015). doi: 10.1038/ni.3069
- Madissoon E, et al. A spatially resolved atlas of the human lung characterizes a gland-associated immune niche. Nat Genet 55: 66–77 (2022). doi: 10.1038/s41588-022-01243-4
- Sealy R, et al. Antibody-secreting cells in respiratory tract tissues in the absence of eosinophils as supportive partners. Int Immunol 28: 559–564 (2016). doi: 10.1093/intimm/dxw035
