Identification of a tumor–specific gene regulatory network in human B-cell lymphoma
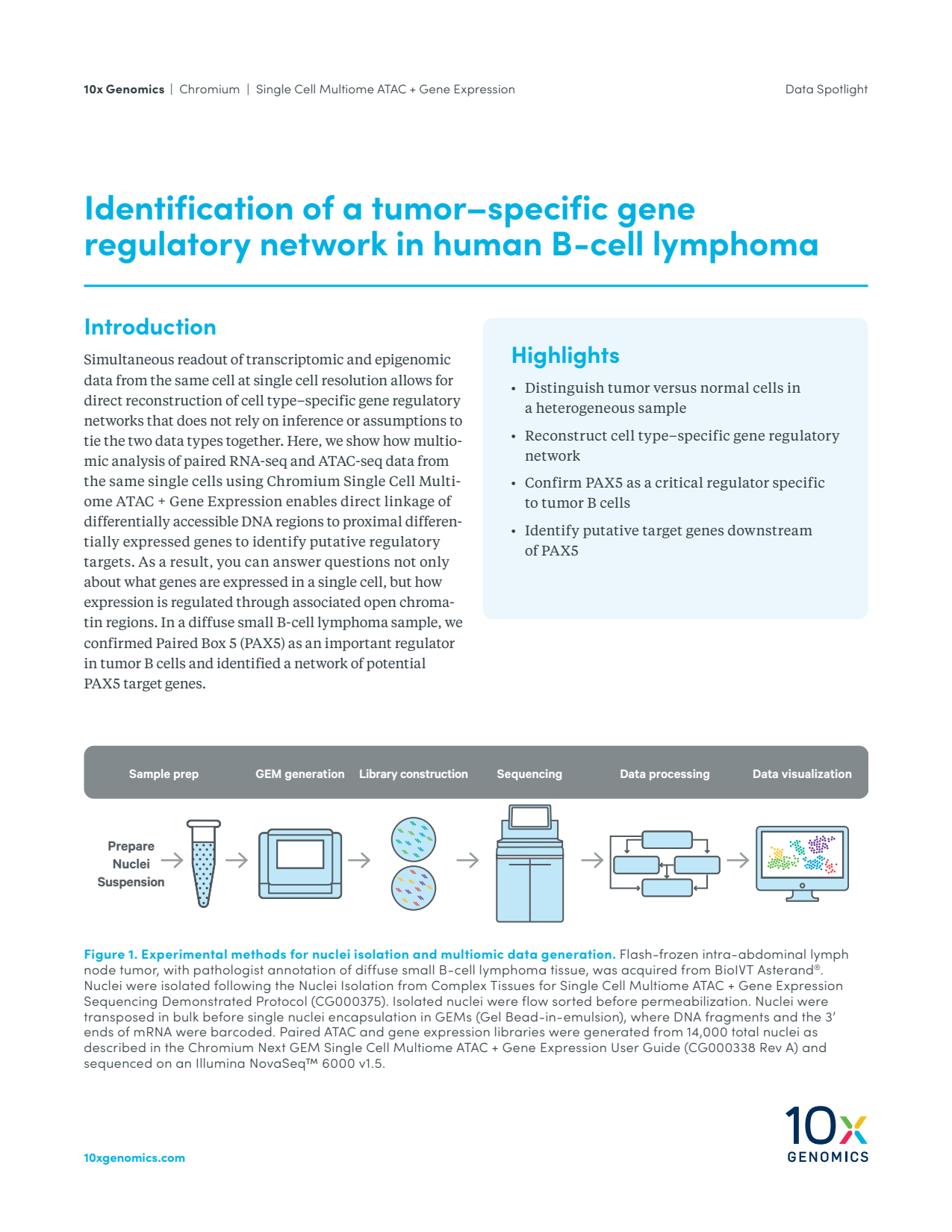
Simultaneous readout of transcriptomic and epigenomic data from the same cell at single cell resolution allows for direct reconstruction of cell type–specific gene regulatory networks that does not rely on inference or assumptions to tie the two data types together. Here, we show how multiomic analysis of paired RNA-seq and ATAC-seq data from the same single cells using Chromium Single Cell Multiome ATAC + Gene Expression enables direct linkage of differentially accessible DNA regions to proximal differentially expressed genes to identify putative targets.
Simultaneous readout of transcriptomic and epigenomic data from the same cell at single cell resolution allows for direct reconstruction of cell type–specific gene regulatory networks that does not rely on inference or assumptions to tie the two data types together. Here, we show how multiomic analysis of paired RNA-seq and ATAC-seq data from the same single cells using Chromium Single Cell Multiome ATAC + Gene Expression enables direct linkage of differentially accessible DNA regions to proximal differentially expressed genes to identify putative targets.