Key Metrics
- Species and tissueHuman breast
- Preservation methodFFPE
- Off-the-shelf panel usedXenium Human Breast Gene Expression Panel (280 genes)
- Number of custom genes33
- Tissue area0.4 cm²
- Number of cells detected*167,780
- Total transcripts detected*34,442,716
- Median transcripts per cell*166
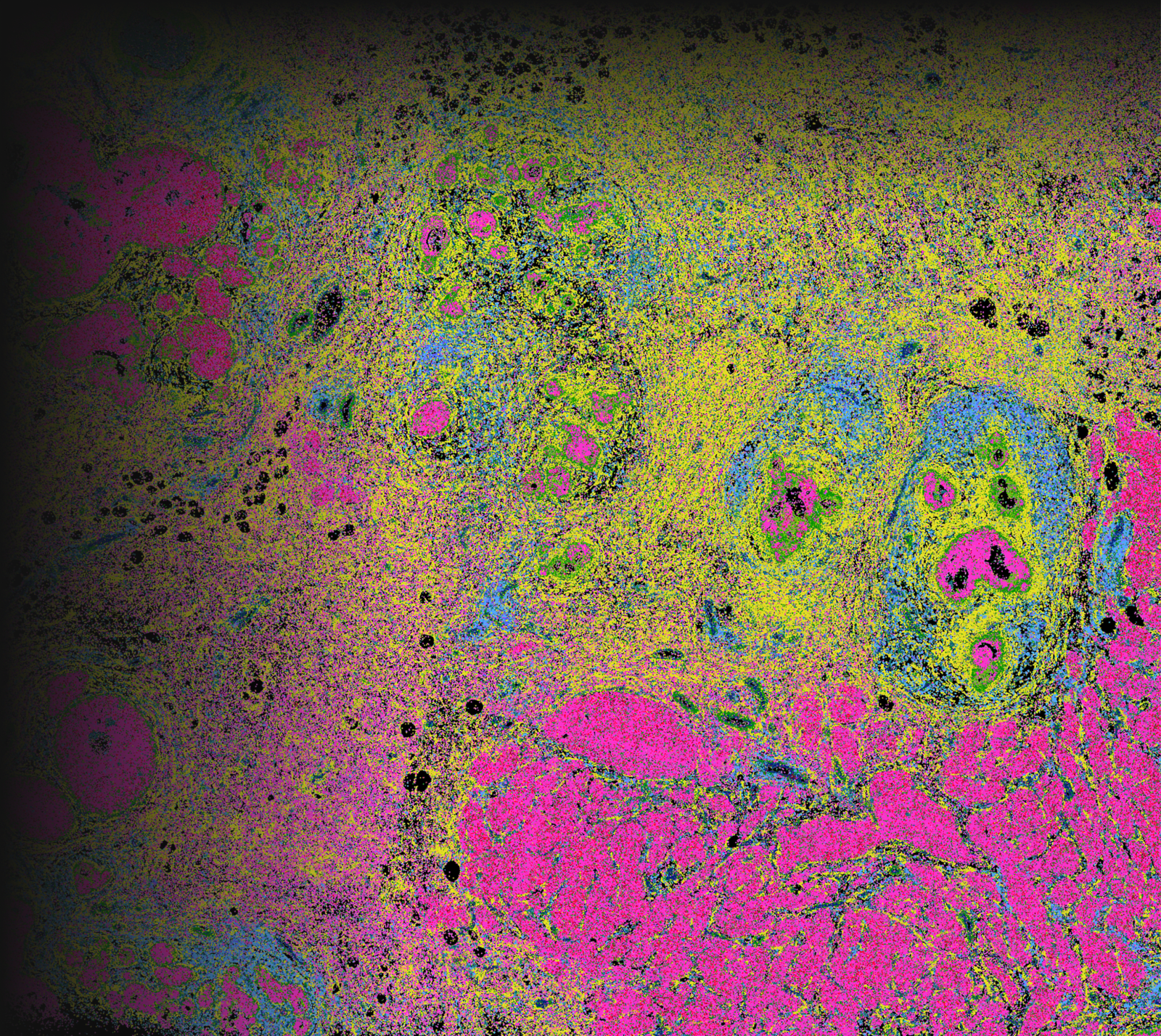
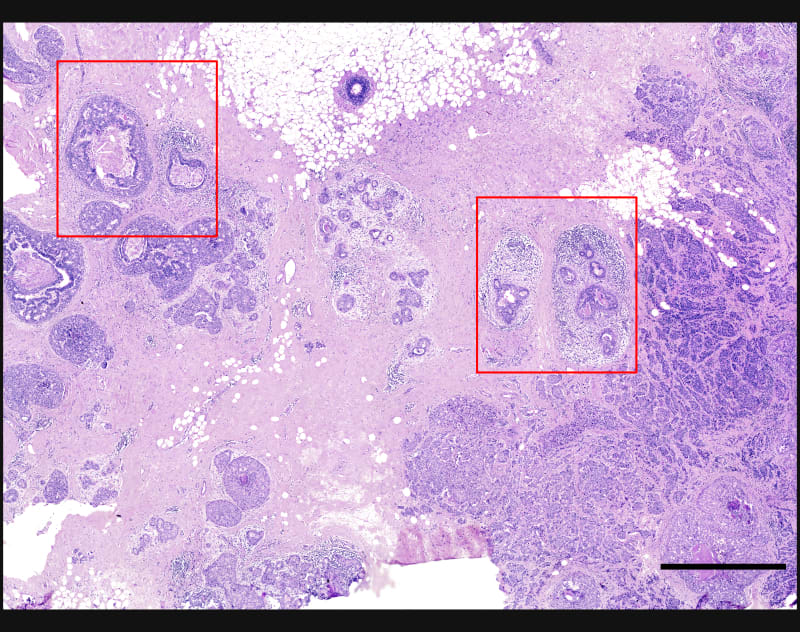
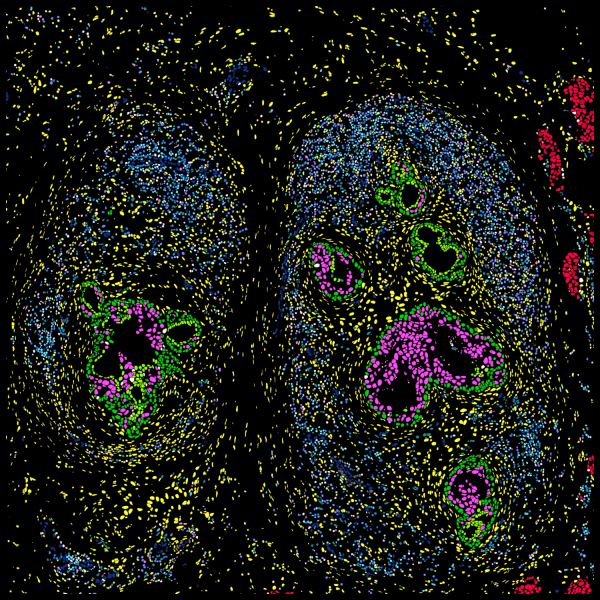
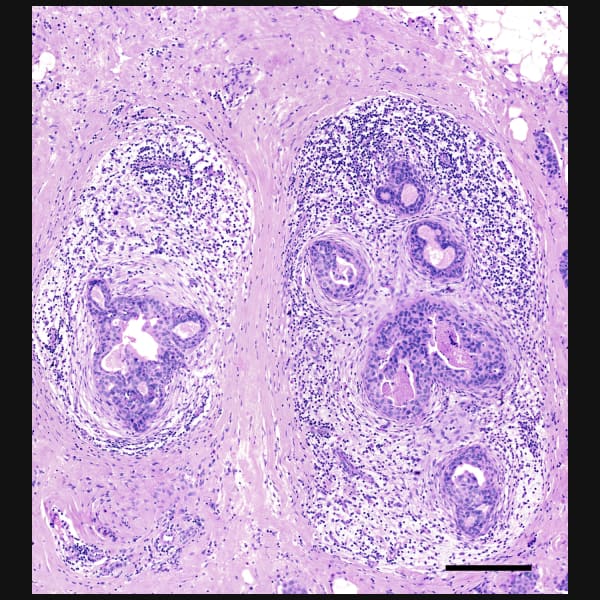
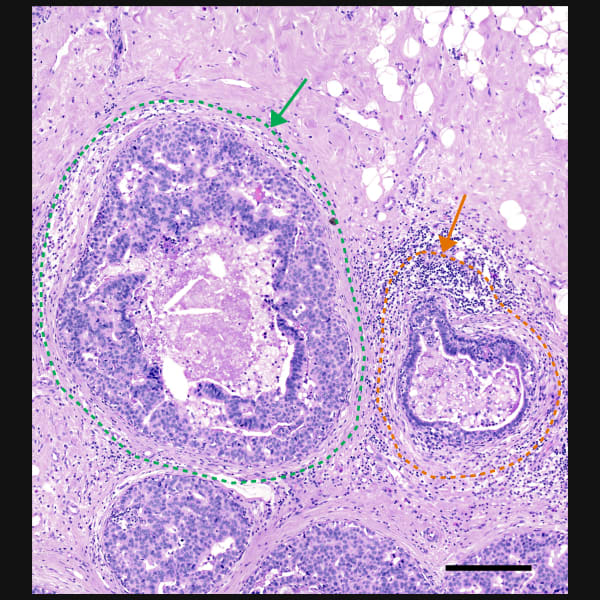
Explore cell type heterogeneity with Xenium and verify with H&E staining on the same section.
Different tissue macrostructures are revealed by the spatial organization of cell types and gene features, capturing vast cellular and molecular heterogeneity within tumor regions
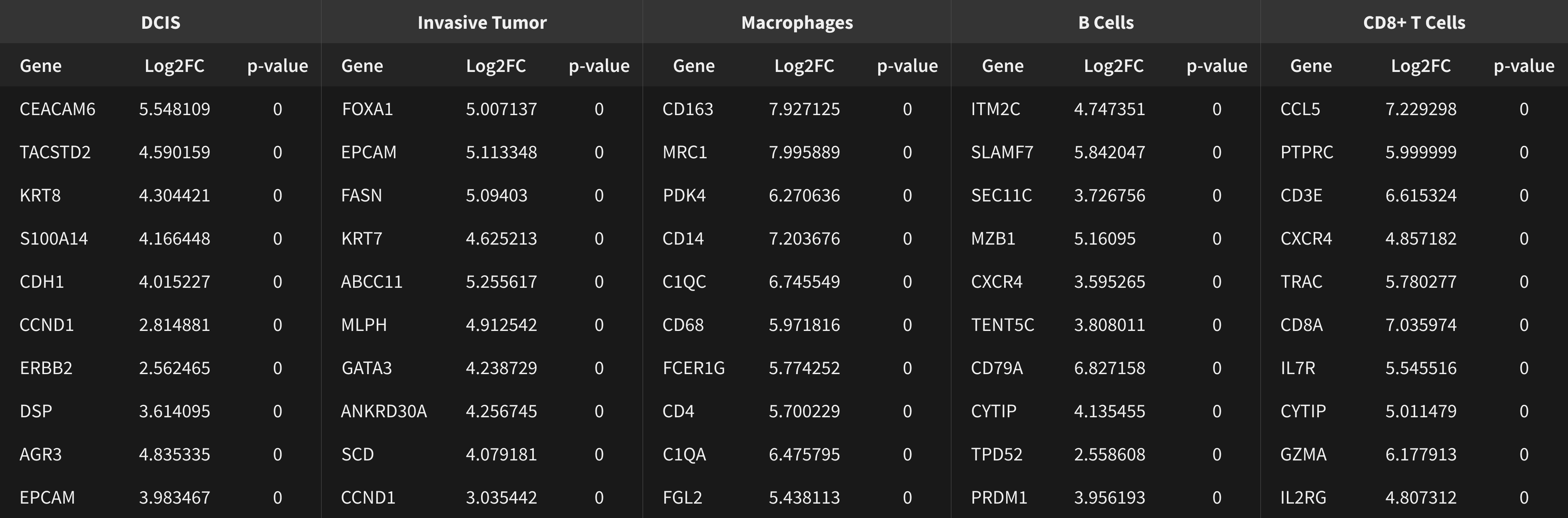
Xenium identifies major cell types in the human breast. As shown in two examples, spatial information is necessary to study tumor subtype heterogeneity and the microenvironment surrounding tumor epithelial cells. Interpretation and validation of rich transcript information is assisted by H&E staining acquired post-Xenium.
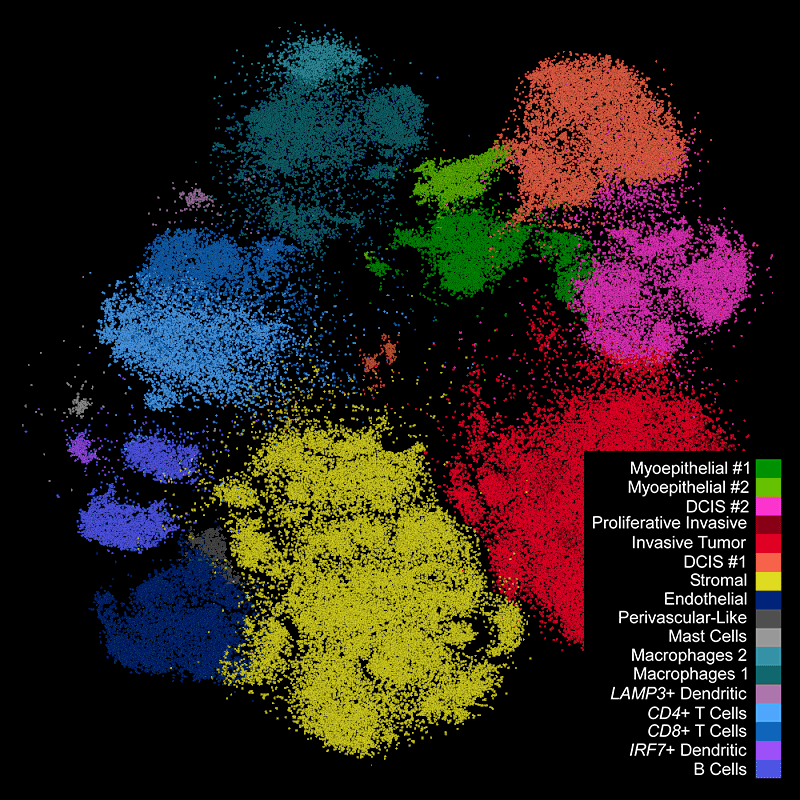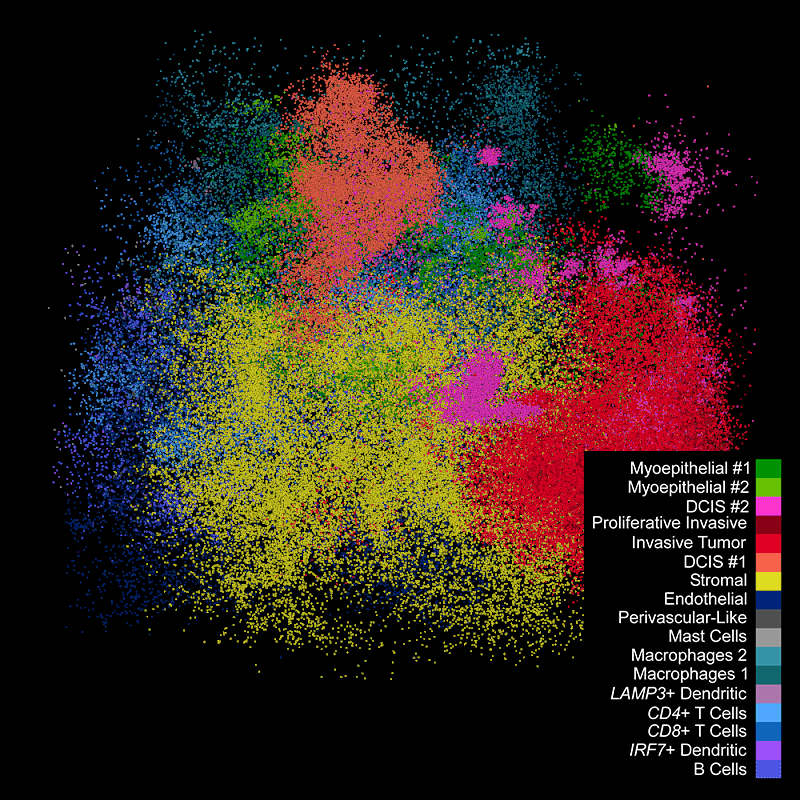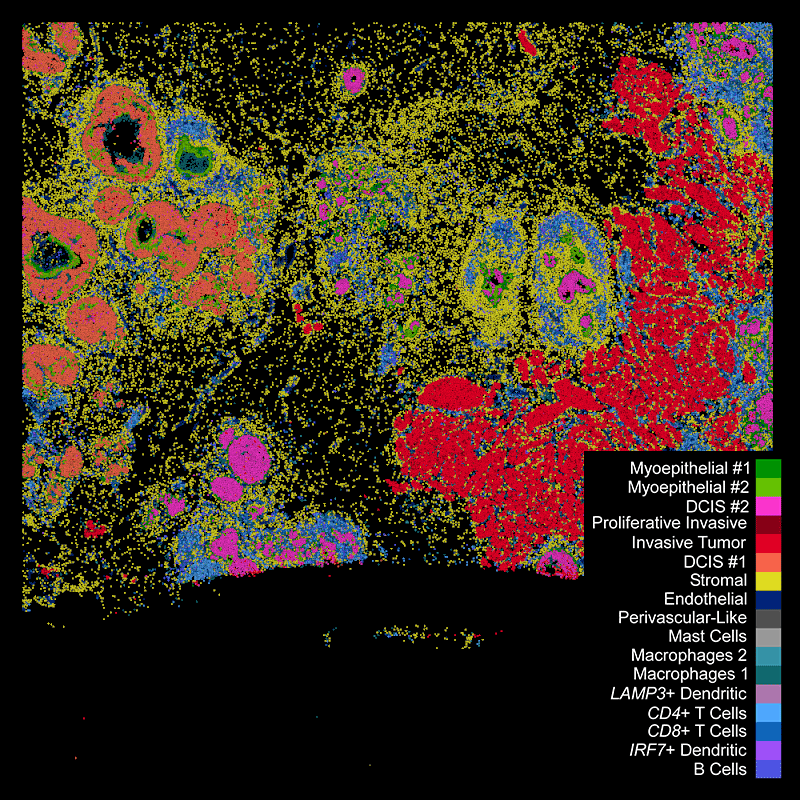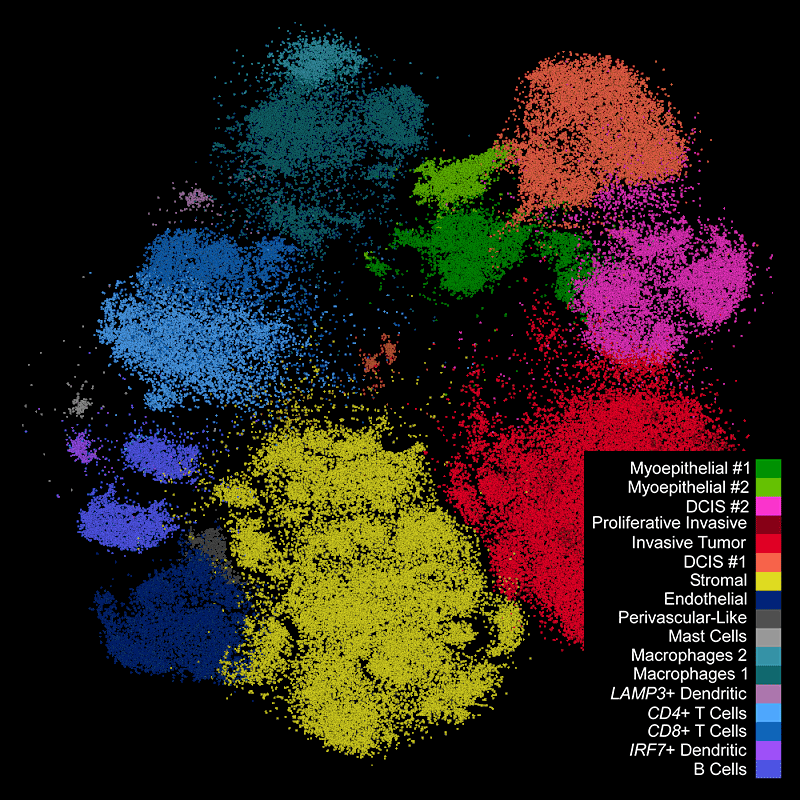

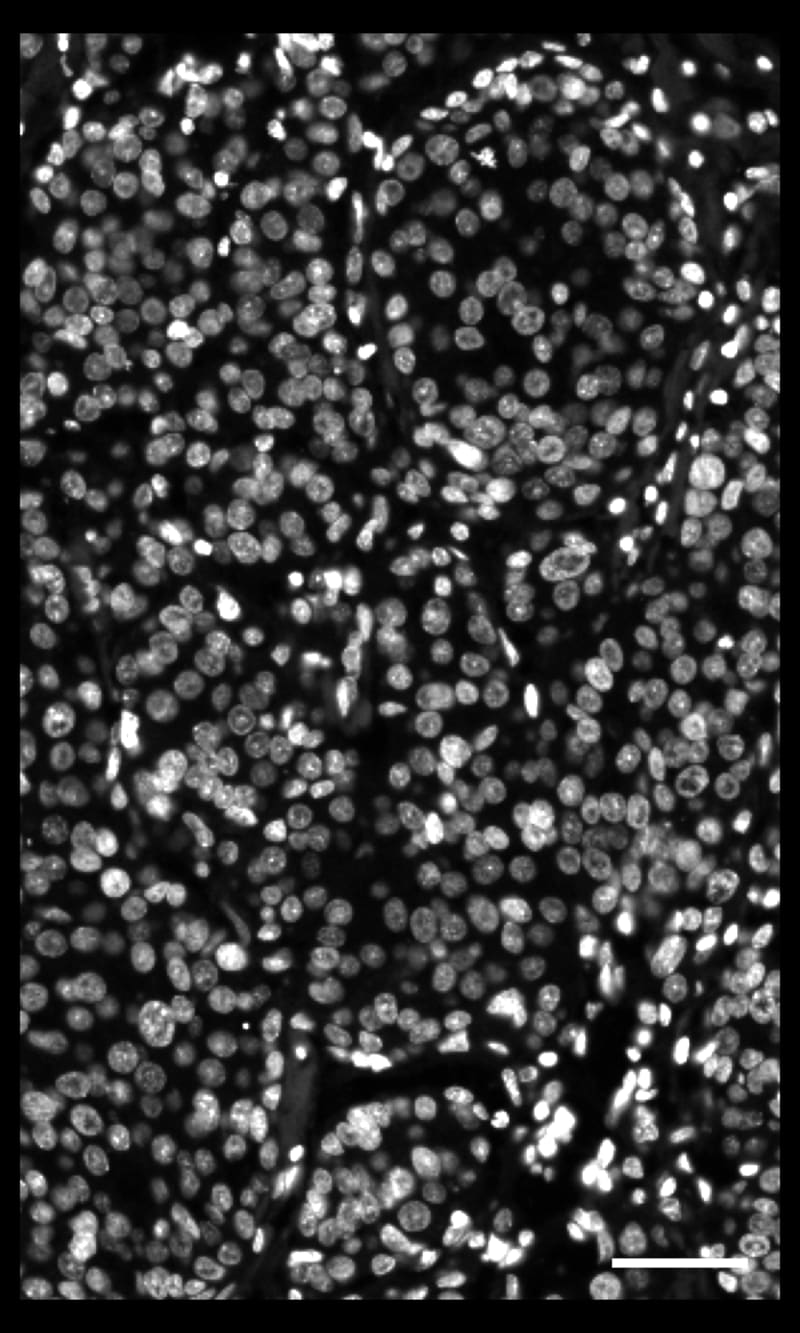
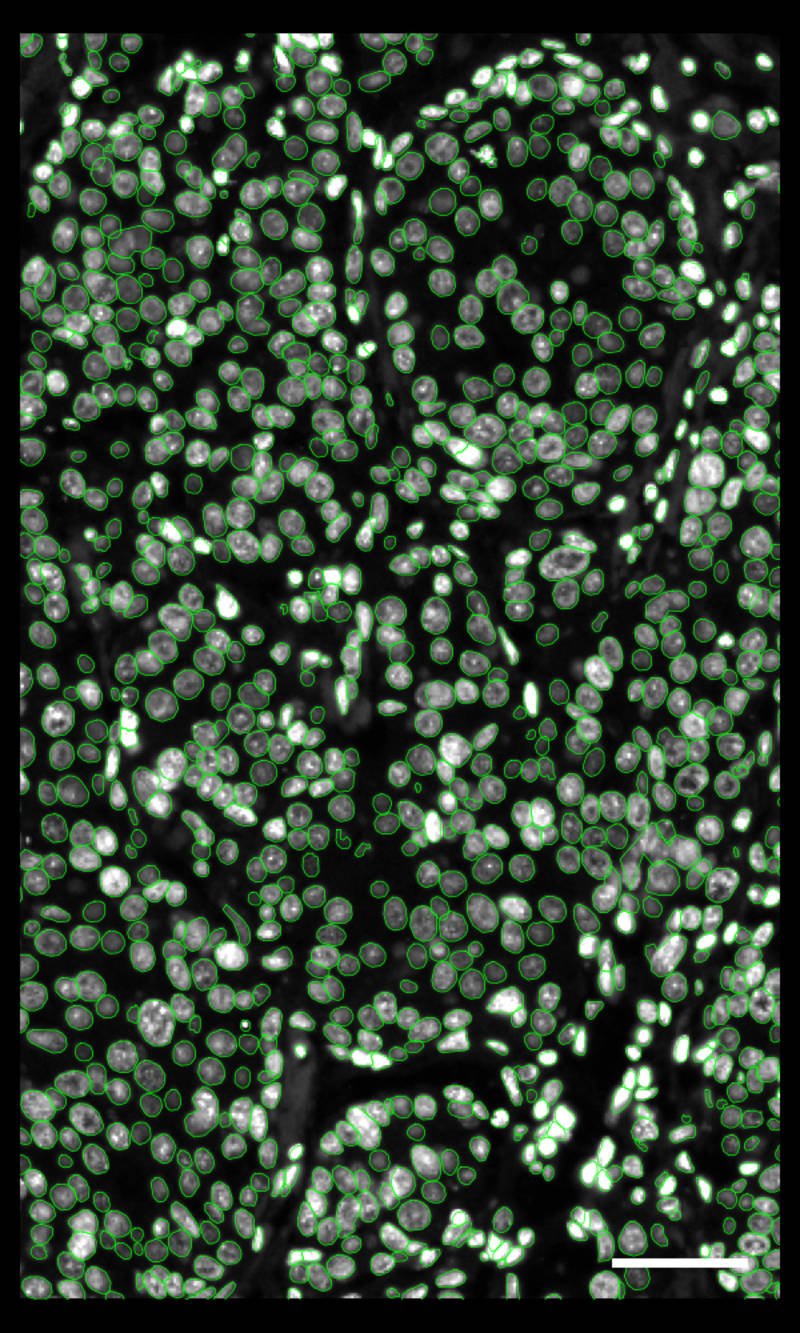
DAPI-based cell segmentation
Xenium uses an in-house trained model to segment nuclei from the DAPI image. Nuclei of irregular shapes are segmented well using Xenium analysis software. Transcripts are assigned based on expansion from nuclei.
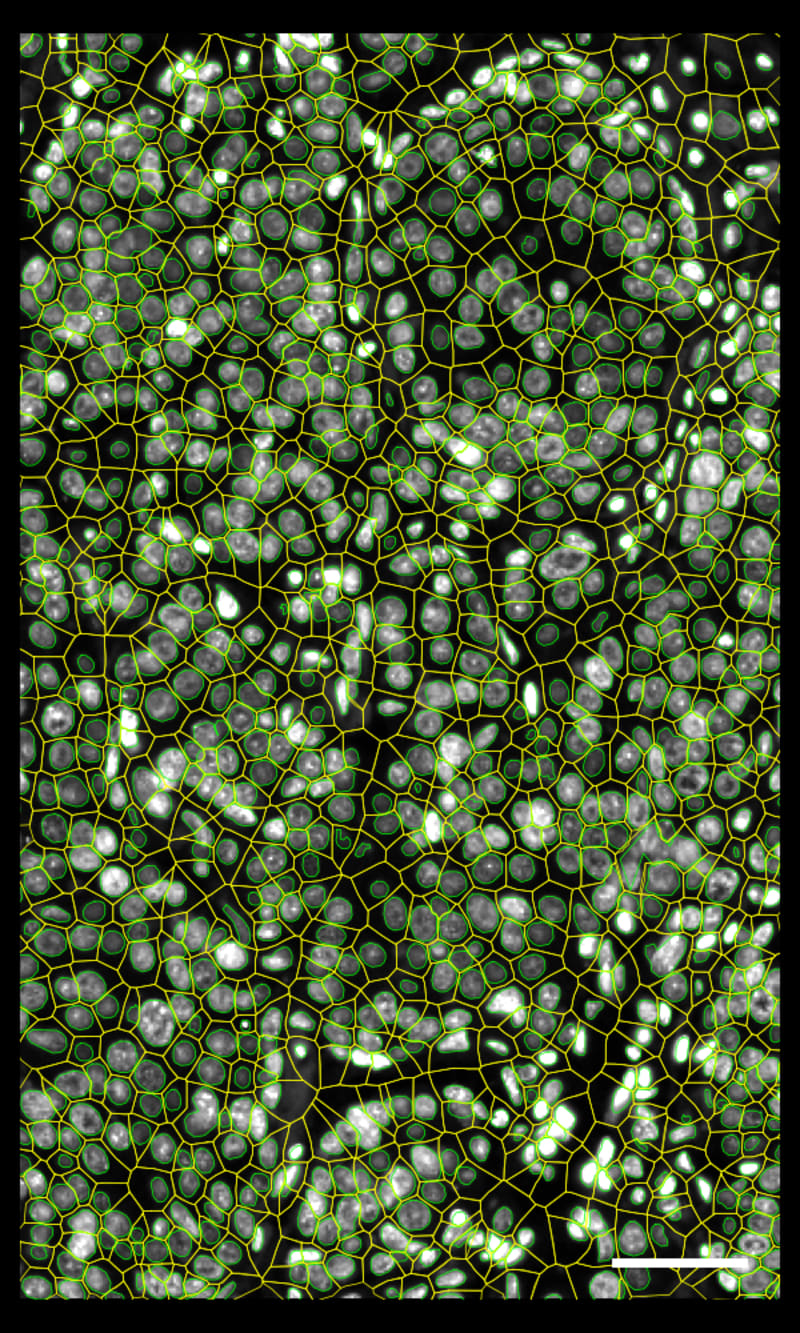
Xenium output can be analyzed in commonly used single cell data analysis software to annotate clusters and/or identify differentially expressed genes.
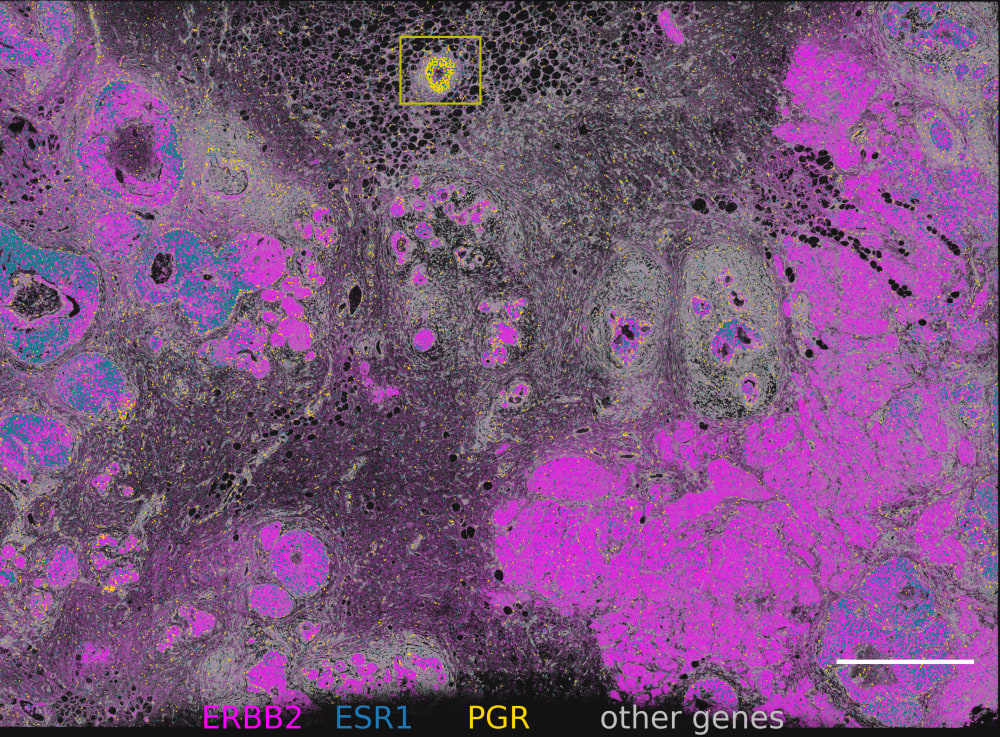
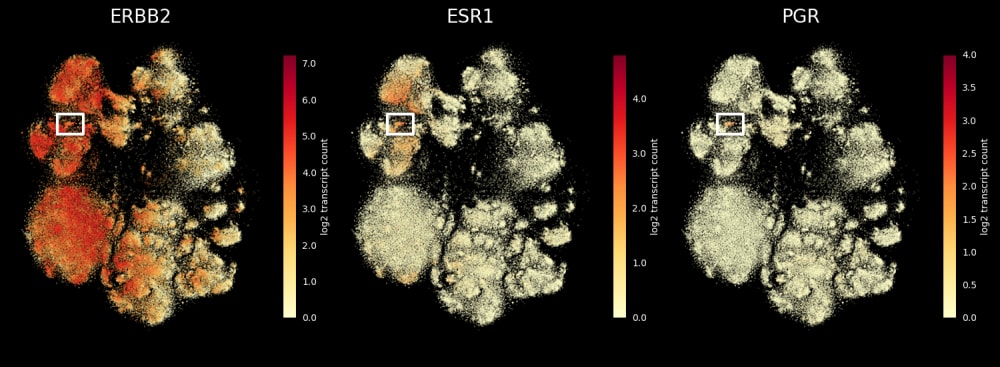
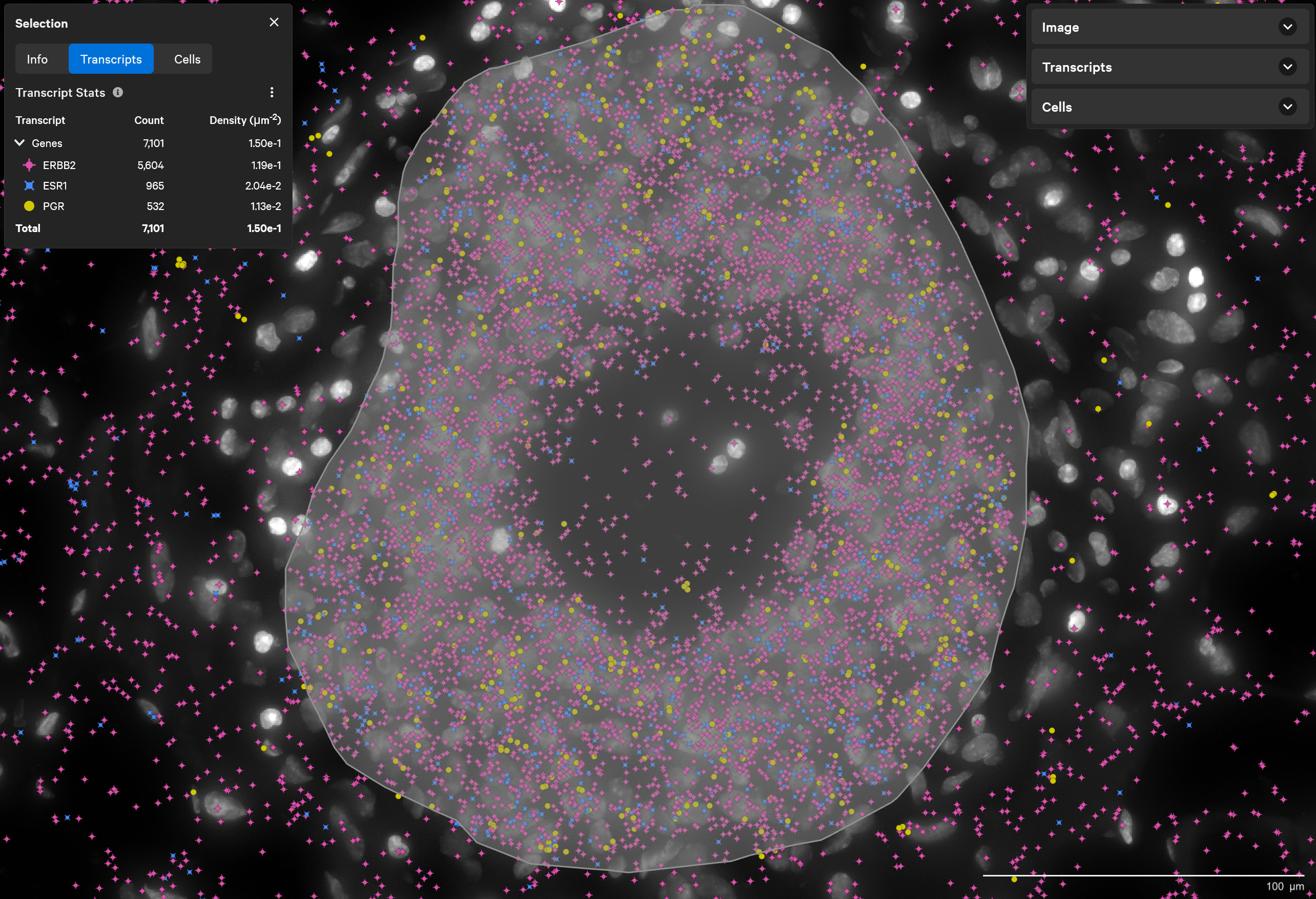
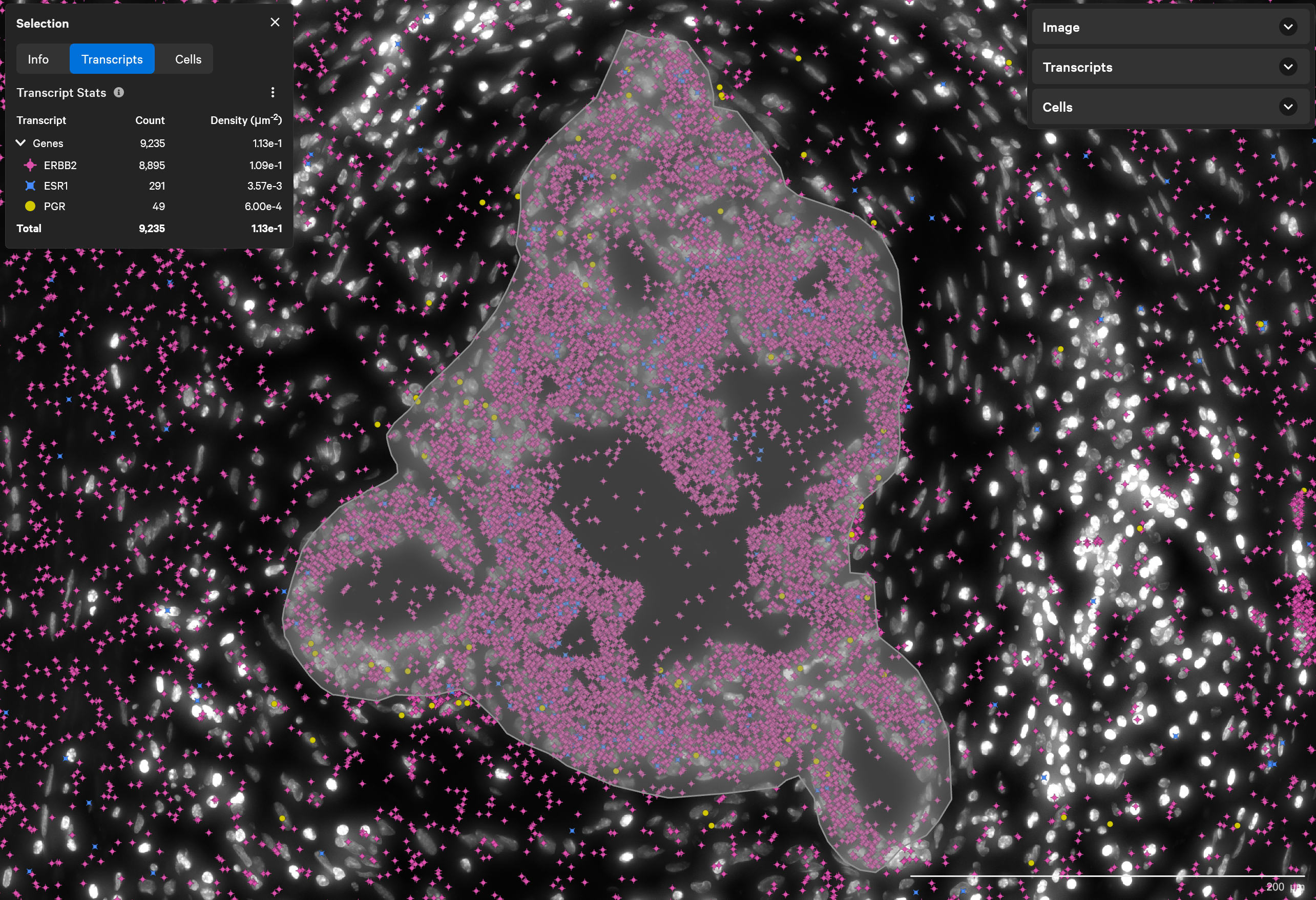
Find triple-positive rare cells
In what is classified as ERBB2(HER2)-positive, ESR1(ER)-positive and PGR(PR)-negative breast tumor, Xenium finds rare triple-positive cells that express all three hormonal receptors.
Explore other Xenium datasets
- High-performance in situ gene expression mapping
- Expertly curated and easily customizable content
- Easy-to-use, high-throughput analyzer
- Intuitive visualization software