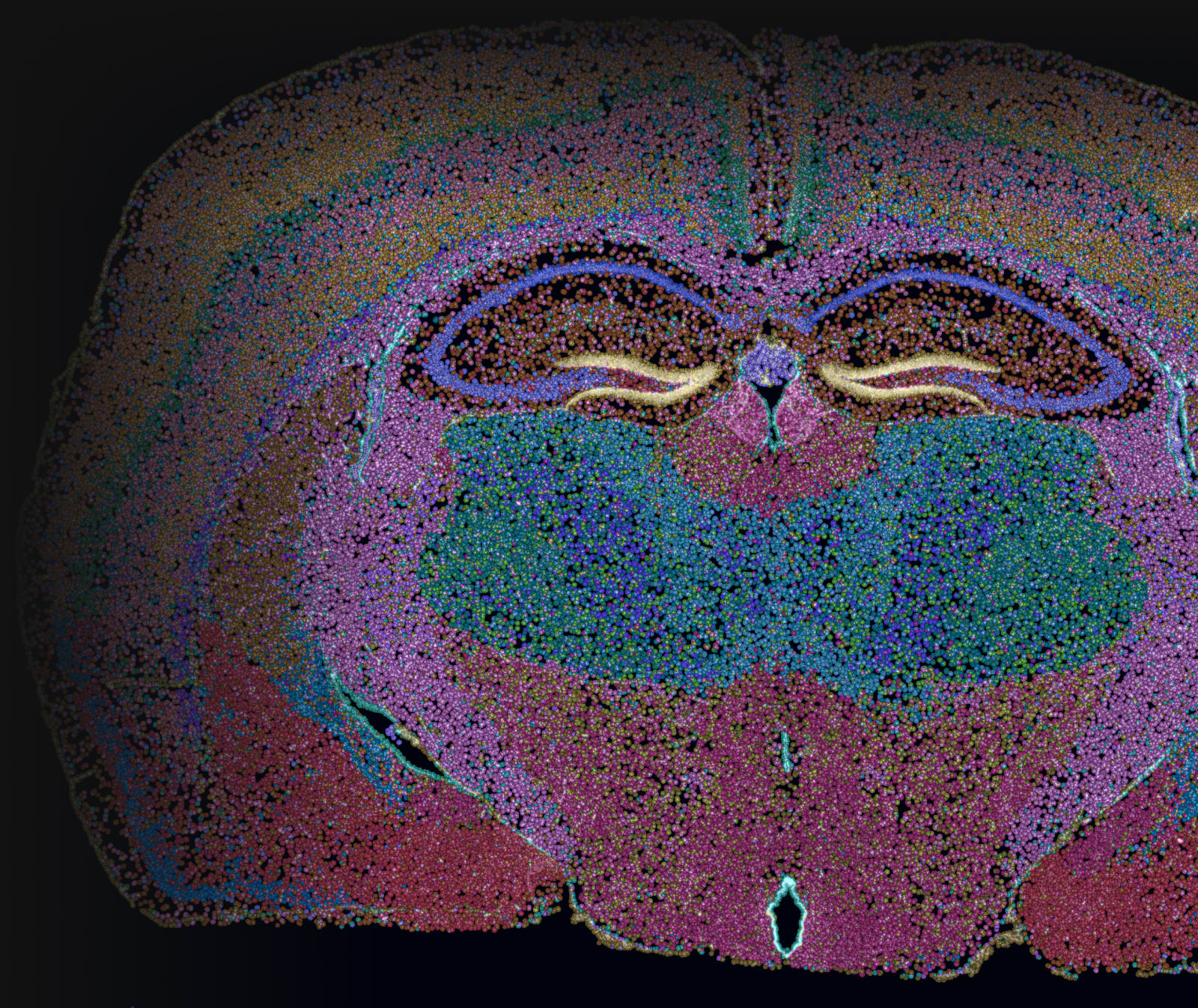
Building subcellular spatial gene expression maps with high sensitivity, specificity, and reproducibility at scale.
Demonstration of Xenium’s high reproducibility across samples
In situ gene expression data acquired from adjacent fresh frozen mouse brain sections placed on the same slide shows low technical variation between replicates.
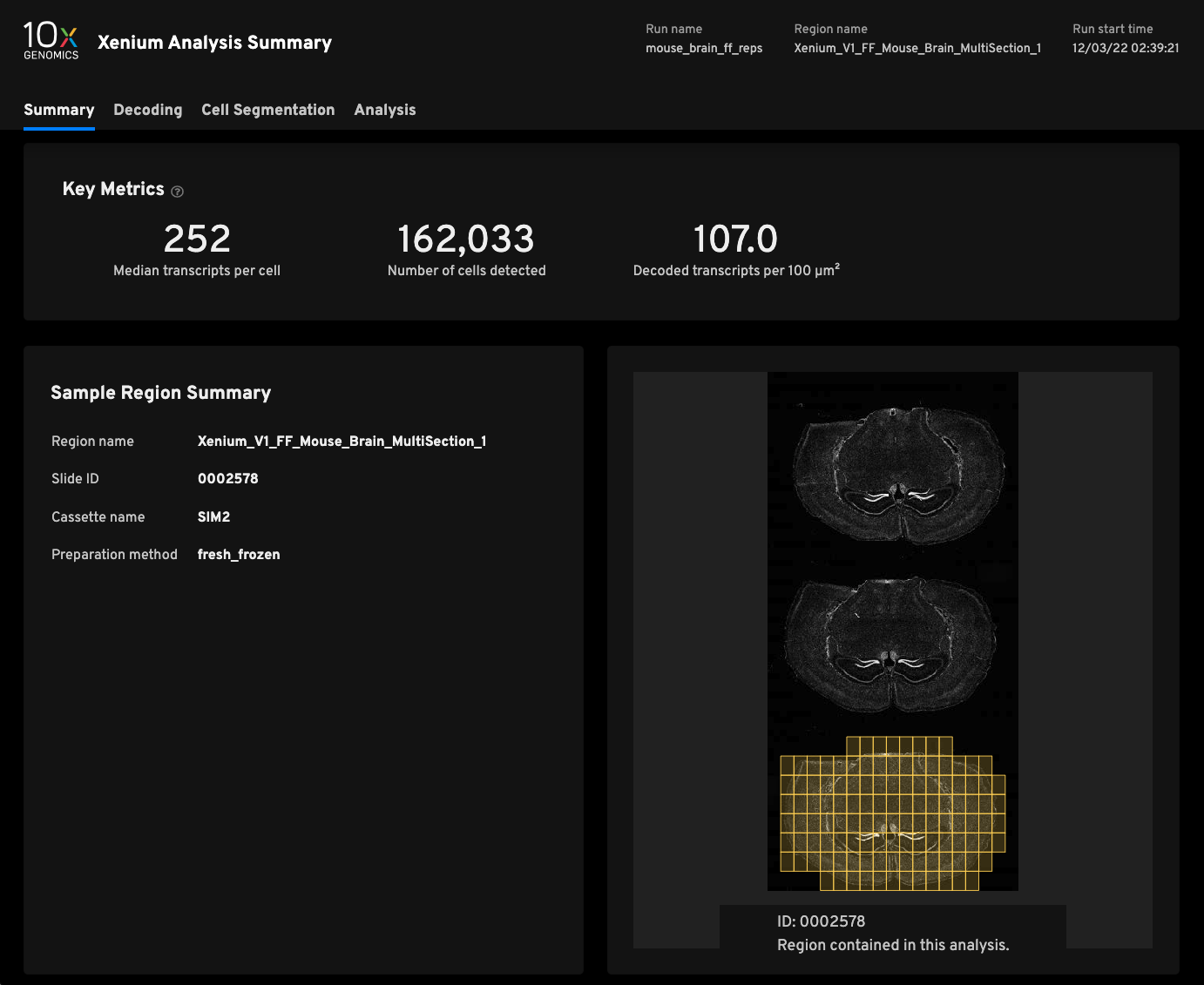
Replicate 1
- Median transcripts per cell252
- Cells detected162,033
- Decoded transcripts per 100μm²107.0
- Total transcripts detected49,587,802
- Scanned tissue area0.63cm²
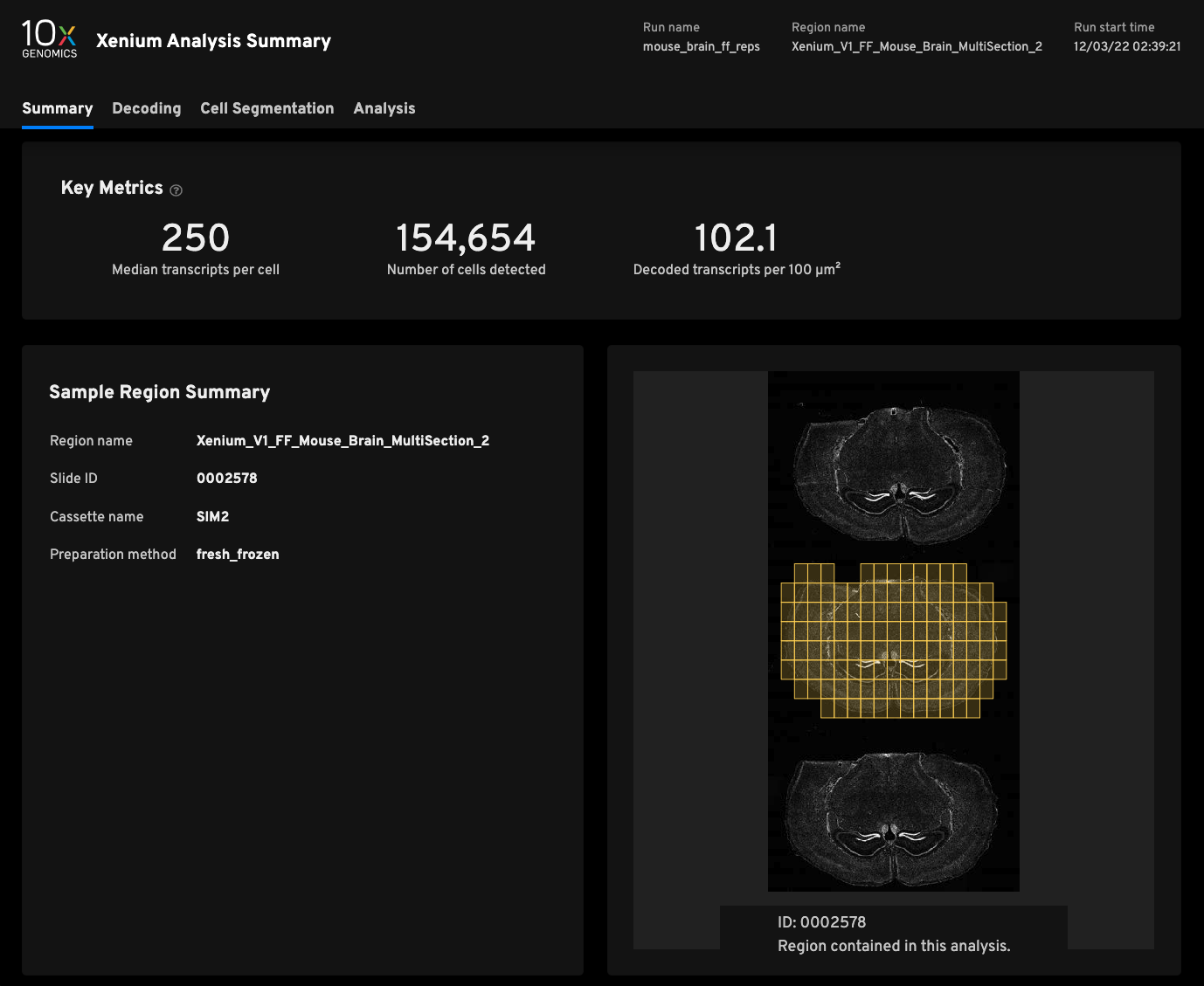
Replicate 2
- Median transcripts per cell250
- Cells detected154,654
- Decoded transcripts per 100μm²102.1
- Total transcripts detected47,093,471
- Scanned tissue area0.64cm²
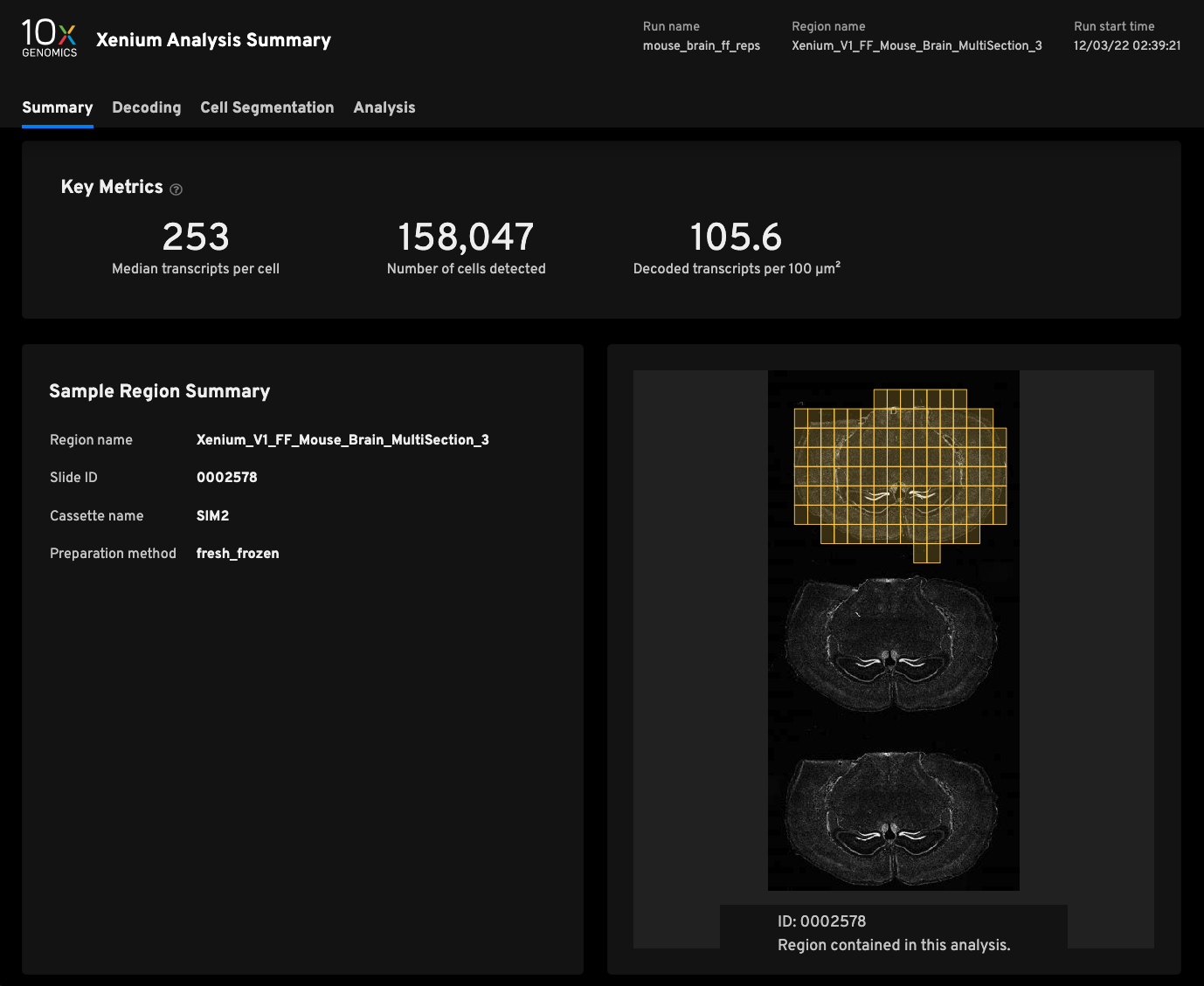
Replicate 3
- Median transcripts per cell253
- Cells detected158,047
- Decoded transcripts per 100μm²105.6
- Total transcripts detected48,460,389
- Scanned tissue area0.61cm²
*Click here to view or download outputs of the dataset from the Xenium Explorer Demo. The tiny dataset is a cropped subset of the full coronal section and has a smaller file size to facilitate creation of data analysis tutorials and vignettes.We recommend reviewing the full coronal sections from the three datasets above for a more detailed representation of the data quality.
Explore other Xenium datasets
Human Breast Cancer
Use Xenium Explorer Demo to explore a Xenium In Situ dataset from a 0.4 cm² human breast cancer FFPE section using a 280-gene Xenium Human Breast Gene Expression Panel with 33 additional custom genes.
Learn about the human breast cancer demo
Xenium In Situ
- High-performance in situ gene expression mapping
- Expertly curated and easily customizable content
- Easy-to-use, high-throughput analyzer
- Intuitive visualization software
Learn about the platform